Epigenetic Reprogramming Drives Epithelial Disruption in Chronic Obstructive Pulmonary Disease
- PMID: 37976469
- PMCID: PMC10914773
- DOI: 10.1165/rcmb.2023-0147OC
Epigenetic Reprogramming Drives Epithelial Disruption in Chronic Obstructive Pulmonary Disease
Abstract
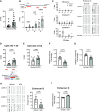
Chronic obstructive pulmonary disease (COPD) remains a major public health challenge that contributes greatly to mortality and morbidity worldwide. Although it has long been recognized that the epithelium is altered in COPD, there has been little focus on targeting it to modify the disease course. Therefore, mechanisms that disrupt epithelial cell function in patients with COPD are poorly understood. In this study, we sought to determine whether epigenetic reprogramming of the cell-cell adhesion molecule E-cadherin, encoded by the CDH1 gene, disrupts epithelial integrity. By reducing these epigenetic marks, we can restore epithelial integrity and rescue alveolar airspace destruction. We used differentiated normal and COPD-derived primary human airway epithelial cells, genetically manipulated mouse tracheal epithelial cells, and mouse and human precision-cut lung slices to assess the effects of epigenetic reprogramming. We show that the loss of CDH1 in COPD is due to increased DNA methylation site at the CDH1 enhancer D through the downregulation of the ten-eleven translocase methylcytosine dioxygenase (TET) enzyme TET1. Increased DNA methylation at the enhancer D region decreases the enrichment of RNA polymerase II binding. Remarkably, treatment of human precision-cut slices derived from patients with COPD with the DNA demethylation agent 5-aza-2'-deoxycytidine decreased cell damage and reduced air space enlargement in the diseased tissue. Here, we present a novel mechanism that targets epigenetic modifications to reverse the tissue remodeling in human COPD lungs and serves as a proof of concept for developing a disease-modifying target.
Keywords: CDH1; COPD; DNA methylation; E-cadherin; epigenetic.
Figures






Comment in
-
Disinherit your Descendants by Rewriting the Chronic Pulmonary Obstructive Disease Epigenetic Script.Am J Respir Cell Mol Biol. 2024 Mar;70(3):153-154. doi: 10.1165/rcmb.2023-0387ED. Am J Respir Cell Mol Biol. 2024. PMID: 38060824 Free PMC article. No abstract available.
References
MeSH terms
Substances
Grants and funding
- HHSN268200900019C/GM/NIGMS NIH HHS/United States
- R01HL144718/GM/NIGMS NIH HHS/United States
- P01 HL107202/HL/NHLBI NIH HHS/United States
- HHSN268200900015C/GM/NIGMS NIH HHS/United States
- U01 HL137880/HL/NHLBI NIH HHS/United States
- U01HL137880/GM/NIGMS NIH HHS/United States
- HHSN268200900018C/GM/NIGMS NIH HHS/United States
- HHSN268200900014C/GM/NIGMS NIH HHS/United States
- R01 AI129958/AI/NIAID NIH HHS/United States
- HHSN268200900014C/HL/NHLBI NIH HHS/United States
- S10 OD023548/OD/NIH HHS/United States
- HHSN268200900019C/HL/NHLBI NIH HHS/United States
- HHSN268200900020C/GM/NIGMS NIH HHS/United States
- U24 HL141762/HL/NHLBI NIH HHS/United States
- U24HL141762/GM/NIGMS NIH HHS/United States
- K24 HL137013/HL/NHLBI NIH HHS/United States
- U01 ES026721/ES/NIEHS NIH HHS/United States
- R01 HL124099/HL/NHLBI NIH HHS/United States
- HHSN268200900015C/HL/NHLBI NIH HHS/United States
- 5U01ES026721/ES/NIEHS NIH HHS/United States
- HHSN268200900016C/HL/NHLBI NIH HHS/United States
- HHSN268200900013C/GM/NIGMS NIH HHS/United States
- R01 HL151107/HL/NHLBI NIH HHS/United States
- U19 AI077439/AI/NIAID NIH HHS/United States
- R01HL124099/HL/NHLBI NIH HHS/United States
- R01HL182622/GM/NIGMS NIH HHS/United States
- HHSN268200900018C/HL/NHLBI NIH HHS/United States
- HHSN268200900017C/HL/NHLBI NIH HHS/United States
- HHSN268200900020C/HL/NHLBI NIH HHS/United States
- HHSN268200900013C/HL/NHLBI NIH HHS/United States
- HLR01HL151107/HL/NHLBI NIH HHS/United States
- HHSN268200900016C/GM/NIGMS NIH HHS/United States
- R01 HL144718/HL/NHLBI NIH HHS/United States
- S10OD023548/GM/NIGMS NIH HHS/United States
- HHSN268200900017C/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

