CD_99 G1 neutrophils modulate osteogenic differentiation of mesenchymal stem cells in the pathological process of ankylosing spondylitis
- PMID: 37977819
- PMCID: PMC10894850
- DOI: 10.1136/ard-2023-224107
CD_99 G1 neutrophils modulate osteogenic differentiation of mesenchymal stem cells in the pathological process of ankylosing spondylitis
Abstract
Objectives: This study aimed to identify the types and heterogeneity of cells within the spinal enthesis and investigate the underlying mechanisms of osteogenesis.
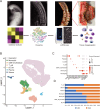
Methods: Single-cell RNA sequencing was used to identify cell populations and their gene signatures in the spinal enthesis of five patients with ankylosing spondylitis (AS) and three healthy individuals. The transcriptomes of 40 065 single cells were profiled and divided into 7 clusters: neutrophils, monocytic cells, granulomonocytic progenitor_erythroblasts, T cells, B cells, plasma cells and stromal cells. Real-time quantitative PCR, immunofluorescence, flow cytometry, osteogenesis induction, alizarin red staining, immunohistochemistry, short hairpin RNA and H&E staining were applied to validate the bioinformatics analysis.
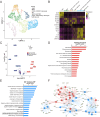
Results: Pseudo-time analysis showed two differentiation directions of stromal cells from the mesenchymal stem cell subpopulation MSC-C2 to two Cxcl12-abundant-reticular (CAR) cell subsets, Osteo-CAR and Adipo-CAR, within which three transcription factors, C-JUN, C-FOS and CAVIN1, were highly expressed in AS and regulated the osteogenesis of mesenchymal stem cells. A novel subcluster of early-stage neutrophils, CD99_G1, was elevated in AS. The proinflammatory characteristics of monocyte dendritic cell progenitor-recombinant adiponectin receptor 2 monocytic cells were explored. Interactions between Adipo-CAR cells, CD99_G1 neutrophils and other cell types were mapped by identifying ligand-receptor pairs, revealing the recruitment characteristics of CD99_G1 neutrophils by Adipo-CAR cells and the pathogenesis of osteogenesis induced in AS.
Conclusions: Our results revealed the dynamics of cell subpopulations, gene expression and intercellular interactions during AS pathogenesis. These findings provide new insights into the cellular and molecular mechanisms of osteogenesis and will benefit the development of novel therapeutic strategies.
Keywords: Ankylosing Spondylitis; Autoimmune Diseases; Inflammation.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

