Identification of gene expression biomarkers to predict clinical response to methotrexate in patients with rheumatoid arthritis
- PMID: 37978145
- PMCID: PMC10774206
- DOI: 10.1007/s10067-023-06814-2
Identification of gene expression biomarkers to predict clinical response to methotrexate in patients with rheumatoid arthritis
Abstract
Objectives: To identify biomarkers at the gene expression level to predict response to methotrexate (MTX) in patients with rheumatoid arthritis (RA).
Methods: MTX-naïve patients with RA were started on MTX and followed up over three months. The disease activity score 28 (DAS28) was used to classify patients into responders and non-responders. Genome-wide gene expression analysis was performed in CD4 + and CD14 + mononuclear cells sampled from whole blood at baseline to identify differentially expressed genes in responders versus non-responders. Gene selection methods and prediction modelling obtained the most relevant differentially expressed genes. A logistic regression prediction model was subsequently constructed and validated via bootstrapping. The area under the receiver operating characteristic (AUC) curve was calculated to judge model quality.
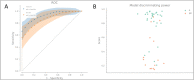
Results: Seventy-nine patients with RA (53.4 ± 13.9 years, 74.7% females) were enrolled, and 70 finished the study with a documented treatment EULAR response (77.1% responders). Forty-six differentially expressed genes were found. The most promising genes were KRTAP4-11, LOC101927584, and PECAM1 in CD4 + cells and PSMD5 and ID1 in CD14 + cells. The final prediction model using these genes reached an AUC of 90%; the validation set's AUC was 82%.
Conclusions: Our prediction model constructed via genome-wide gene expression analysis in CD4 + and CD14 + mononuclear cells yielded excellent predictions. Our findings necessitate confirmation in other cohorts of MTX-naïve RA patients. Especially if used in conjunction with previously identified clinical and laboratory (bio)markers, our results could help predict response to MTX in RA to guide treatment decisions. Key Points • Patients with rheumatoid arthritis may or may not respond to treatment with methotrexate, which is the recommended first-line drug in guidelines around the world. • In non-responders, valuable time is lost until second-line treatments are started. • This study aimed at predicting response to methotrexate by identifying differentially expressed genes from peripheral blood samples. • The final prediction model yielded excellent prognostic values, but validation in other cohorts is necessary to corroborate these findings.
Keywords: Biomarkers; Gene signature; Methotrexate; Pharmacogenomics; Prediction; Response; Rheumatoid arthritis.
© 2023. The Author(s).
Conflict of interest statement
FB reports receiving honoraria and grant/study support from medac, Hexal, and Pfizer. MS and JK have received fees for service and consulting from Charite and medac. AB is an employee of medac GmbH. The other authors have nothing to disclose.
Figures

References
-
- Smolen JS, Landewé RBM, Bijlsma JWJ, Burmester GR, Dougados M, Kerschbaumer A, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann Rheum Dis. 2020;79(6):685–699. doi: 10.1136/annrheumdis-2019-216655. - DOI - PubMed
-
- Deutsches Rheumaforschungszentrum: Daten der Kerndokumentation 2019. https://www.drfz.de/wp-content/uploads/ergebnisse-standardprasentation-2... (2019). Accessed January 26th 2022
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

