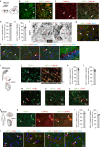
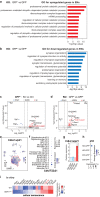
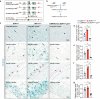
Fig. 4. Selective deletion of GDF11 in the EN causes their own hyperexcitability and deteriorates social cognition and object recognition memory in mice.
a Schematic diagrams (left) and representative images (right) of the cingulate gyrus 2 (Cg2), in the prefrontal cortex of GDF11f/f mice aged 4M-5M, where bilateral focal injection of AAV9-CaMKIIα-Cre-P2A-GFP virus (KO) or AAV9-CaMKIIα-GFP virus (Ctrl) was received at age of 2–3 M and survived for two more months. b Infrared-differential interference contrast (IR-DIC) image (top) and GFP fluorescent image (bottom) of an example of GFP+ EN which is undergoing whole-cell patch clamp recording (n = 64 cells from six mice). c Representative whole-cell recordings in brain slice of a control EN (in Cg2 of GDF11f/f mice, Ctrl, blue) and a GDF11 deleted-EN (in Cg2 of fGDF11cKO mice, KO, red) show the firing of action potentials (AP) in response to a series of step current injections. d Examples show typical firing patterns of GFP+ EN of fGDF11cKO mice. e Pie graphs show the percentage of GFP+ EN with diverse firing patterns (RS, regular spiking; IS, irregular spiking; IB, intrinsic bursting; RB, repetitive bursting) in WT or KO mice. f Left, plots of the AP frequency as a function of injected currents. Curves are color coded (Ctrl, blue, n = 31 cells from three mice; KO, red, n = 33 cells from three mice). Inset shows the beginning of the curve. Right, plots of the rheobase (Ctrl: 113 ± 16 vs. KO: 81 ± 10 pA, P = 0.049) and slope (Ctrl: 0.18 ± 0.01 vs. KO: 0.30 ± 0.03, P = 0.000) in the two groups (Ctrl, n = 31 cells from three mice; KO, n = 30 cells from three mice). g Left, representative AP waveforms (top) and phase plots (bottom) from Ctrl (blue) or KO (red) group. Right, plots of the AP threshold (Ctrl: −37.9 ± 0.8 vs. KO: −35.0 ± 0.7 mV, P = 0.014), amplitude (AMP) (Ctrl: 85.8 ± 1.6 vs. KO: 78.6 ± 2.2 mV, P = 0.010) and half-width (Ctrl: 0.79 ± 0.03 vs. KO: 0.74 ± 0.03 ms, P = 0.30) in the two groups (Ctrl, n = 29 cells from three mice; KO, n = 24 cells from three mice). h Left-top, representative membrane potential responses to negative current pulses from Ctrl (blue) or KO (red) groups. Plots of the input resistance (Ctrl: 104 ± 10 vs. KO: 214 ± 21 MΩ, P = 0.000), membrane constant (Ctrl: 14.4 ± 1.1 vs. KO: 22.1 ± 2.0 ms, P = 0.003), Sag ratio (Ctrl: 1.18 ± 0.02 vs. KO: 1.27 ± 0.03, P = 0.033), membrane capacitance (Ctrl: 147 ± 11 vs. KO: 95 ± 5 pF, P = 0.000) and RMP (Ctrl: −67.3 ± 1.0 vs. KO: −63.1 ± 0.9 mV, P = 0.004) in the two groups (Ctrl, n = 31 cells from three mice; KO, n = 33 cells from three mice). i Representative whole-cell recordings of mIPSC from the EN in GDF11f/f mice (Ctrl, blue) and fGDF11cKO mice (KO, red). j Left, scaled mIPSC examples in the two groups. Right, plots of rising time (Ctrl: 0.65 ± 0.04 vs. KO: 0.85 ± 0.06 ms, P = 0.005) and decay time (Ctrl: 4.44 ± 0.21 vs. KO: 4.69 ± 0.34 ms, P = 0.53) of mIPSCs in the two groups (Ctrl, n = 18 cells from four mice; KO, n = 16 cells from four mice). k, l Cumulative frequency curve of the inter-event-interval (k) and amplitude (l) of mIPSCs. Insets show the group plots of mIPSC frequency (k, Ctrl: 34.6 ± 5.2 vs. KO: 4.0 ± 0.9 Hz, P = 0.000) and amplitude (l, Ctrl: 24.0 ± 1.6 vs. KO: 20.5 ± 1.8 pA, P = 0.16). m–p Recordings of mEPSCs (Ctrl, n = 24 cells from four mice; KO, n = 28 cells from 4 mice) and similar plots as the mIPSCs shown above. Rising time (n, ctrl: 0.87 ± 0.05 vs. KO: 0.81 ± 0.06 ms, P = 0.46); Decay time (n, ctrl: 3.54 ± 0.20 vs. KO: 2.98 ± 0.24 ms, P = 0.041); Frequency (o, Ctrl: 3.66 ± 0.84 vs. KO: 3.13 ± 0.65 Hz, p = 0.82); Amplitude (p, Ctrl: 14.5 ± 0.8 vs. KO: 14.3 ± 0.9 pA, P = 0.33). q, r Representative traces showing IPSC (q, left) or EPSC (r, left) evoked by extracellular electric stimulations for the comparison of paired-pulse ratio (PPR) in GDF11f/f mice (Ctrl, blue) and fGDF11cKO mice (KO, red). Group plots of PPR for IPSC (q, right, Ctrl, n = 7 cells from 3 mice: 0.98 ± 0.07 vs. KO, n = 9 cells from three mice: 1.16 ± 0.20, P = 0.92) and EPSC (r, right, Ctrl, n = 9 cells from 3 mice: 1.38 ± 0.07 vs. KO, n = 6 cells from three mice: 1.26 ± 0.06, P = 0.24). s Track diagrams in the 3-chamber test (3CT) between the fGDF11cKO (KO) and GDF11f/f (Ctrl) mice aged 4–5 M. O object, S1 stranger mouse, S2 new stranger mouse. t Quantification of the exploration time in 3CT (KO, n = 13; Ctrl, n = 13 mice) on objects between the fGDF11cKO (KO) and GDF11f/f (Ctrl) mice aged 4–5 M. O1, object 1; O2, object 2. u Quantification of the preference index (S1-O) between the S1 and object in the KO and Ctrl groups (KO, n = 13; Ctrl, n = 13 mice). v Quantification of the preference index (S2-S1) between the S2 and S1 in the KO and Ctrl groups (KO, n = 13; Ctrl, n = 13 mice). w Schematic diagram of the novel object recognition test (NORT) between the GDF11cKO and GDF11f/f mice aged 10 M. Red squares indicate the familiar toy while blue triangle indicates a novel toy. x Quantification of the percentage of exploration time (GDF11cKO, n = 9; GDF11f/f, n = 6 mice) on the familiar or a novel toy in the GDF11cKO and GDF11f/f mice aged 10 M. y Quantification of the novel object discrimination index ((novel-familiar)/(novel + familiar)) between the familiar or a novel toy in the GDF11cKO and GDF11f/f mice aged 10 M (GDF11cKO, n = 9; GDF11f/f, n = 6 mice). Data are presented as mean ± SEM. Whisker boxplots in (f, h) represent the median and interquartile range; whiskers represent 1.5× interquartile range. *P < 0.05, **P < 0.01 and “ns” represents not significant. f (Rheobase/Slope), h (Input resistance/Membrane constant/Sag ratio/Capacitance), j (Rising time), k, n (Decay time), o–q Mann–Whitney U test. g, h (RMP), j (Decay time), l, n (Rising time), r, u (P = 0.0118), v (P = 0.0128), x (GDF11f/f: Familiar versus Novel, P = 0.0331; GDF11cKO: Familiar versus Novel, P = 0.0188) and y (P = 0.0254), unpaired two-tailed t test. t (Ctrl: O1 versus O2, P = 0.3210; KO: O1 versus O2, P = 0.2200), two-way ANOVA with post Sidak’s multiple comparisons test. Source data are provided with this paper.