Spatially distinct molecular patterns of gene expression in idiopathic pulmonary fibrosis
- PMID: 37978501
- PMCID: PMC10655274
- DOI: 10.1186/s12931-023-02572-6
Spatially distinct molecular patterns of gene expression in idiopathic pulmonary fibrosis
Abstract
Background: Idiopathic pulmonary fibrosis (IPF) is a heterogeneous disease that is pathologically characterized by areas of normal-appearing lung parenchyma, active fibrosis (transition zones including fibroblastic foci) and dense fibrosis. Defining transcriptional differences between these pathologically heterogeneous regions of the IPF lung is critical to understanding the distribution and extent of fibrotic lung disease and identifying potential therapeutic targets. Application of a spatial transcriptomics platform would provide more detailed spatial resolution of transcriptional signals compared to previous single cell or bulk RNA-Seq studies.
Methods: We performed spatial transcriptomics using GeoMx Nanostring Digital Spatial Profiling on formalin-fixed paraffin-embedded (FFPE) tissue from 32 IPF and 12 control subjects and identified 231 regions of interest (ROIs). We compared normal-appearing lung parenchyma and airways between IPF and controls with histologically normal lung tissue, as well as histologically distinct regions within IPF (normal-appearing lung parenchyma, transition zones containing fibroblastic foci, areas of dense fibrosis, and honeycomb epithelium metaplasia).
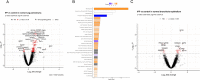
Results: We identified 254 differentially expressed genes (DEGs) between IPF and controls in histologically normal-appearing regions of lung parenchyma; pathway analysis identified disease processes such as EIF2 signaling (important for cap-dependent mRNA translation), epithelial adherens junction signaling, HIF1α signaling, and integrin signaling. Within IPF, we identified 173 DEGs between transition and normal-appearing lung parenchyma and 198 DEGs between dense fibrosis and normal lung parenchyma; pathways dysregulated in both transition and dense fibrotic areas include EIF2 signaling pathway activation (upstream of endoplasmic reticulum (ER) stress proteins ATF4 and CHOP) and wound healing signaling pathway deactivation. Through cell deconvolution of transcriptome data and immunofluorescence staining, we confirmed loss of alveolar parenchymal signals (AGER, SFTPB, SFTPC), gain of secretory cell markers (SCGB3A2, MUC5B) as well as dysregulation of the upstream regulator ATF4, in histologically normal-appearing tissue in IPF.
Conclusions: Our findings demonstrate that histologically normal-appearing regions from the IPF lung are transcriptionally distinct when compared to similar lung tissue from controls with histologically normal lung tissue, and that transition zones and areas of dense fibrosis within the IPF lung demonstrate activation of ER stress and deactivation of wound healing pathways.
© 2023. The Author(s).
Conflict of interest statement
Dr. Blumhagen is supported by sponsored research agreement from Eleven P15, Inc., outside the submitted work. Dr. Schwartz is the founder and unpaid chief scientific officer of Eleven P15, Inc., and serves as a consultant for Vertex Pharmaceuticals. Dr. Fingerlin and Dr. Yang report consulting fees from Eleven P15, Inc., outside the submitted work and Drs. Schwartz, Fingerlin, and Yang have a patent Methods and Compositions for Risk Prediction, Diagnosis, Prognosis, and Treatment of Pulmonary Disorders issued. Dr. Cool reports consultant fees from Eleven P15, Inc. and Theralink Technologies. Dr. Kurche, Avram Walts, and David Heinz report no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
- R01 HL158668/HL/NHLBI NIH HHS/United States
- R01-HL158668/HL/NHLBI NIH HHS/United States
- UG3-HL151865/HL/NHLBI NIH HHS/United States
- UG3 HL151865/HL/NHLBI NIH HHS/United States
- P01 HL162607/HL/NHLBI NIH HHS/United States
- R01 HL149836/HL/NHLBI NIH HHS/United States
- P01 HL092870/HL/NHLBI NIH HHS/United States
- UH3- HL123442/HL/NHLBI NIH HHS/United States
- I01 BX005295/BX/BLRD VA/United States
- R01-HL097163/HL/NHLBI NIH HHS/United States
- P01-HL092870/HL/NHLBI NIH HHS/United States
- R01-HL148437/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

