Comprehensive genomic characterization of HER2-low and HER2-0 breast cancer
- PMID: 37980405
- PMCID: PMC10657399
- DOI: 10.1038/s41467-023-43324-w
Comprehensive genomic characterization of HER2-low and HER2-0 breast cancer
Erratum in
-
Author Correction: Comprehensive genomic characterization of HER2-low and HER2-0 breast cancer.Nat Commun. 2023 Dec 14;14(1):8321. doi: 10.1038/s41467-023-44124-y. Nat Commun. 2023. PMID: 38097580 Free PMC article. No abstract available.
Abstract
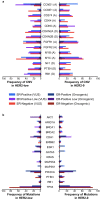
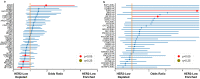
The molecular underpinnings of HER2-low and HER2-0 (IHC 0) breast tumors remain poorly defined. Using genomic findings from 1039 patients with HER2-negative metastatic breast cancer undergoing next-generation sequencing from 7/2013-12/2020, we compare results between HER2-low (n = 487, 47%) and HER2-0 tumors (n = 552, 53%). A significantly higher number of ERBB2 alleles (median copy count: 2.05) are observed among HER2-low tumors compared to HER2-0 (median copy count: 1.79; P = 2.36e-6), with HER2-0 tumors harboring a higher rate of ERBB2 hemideletions (31.1% vs. 14.5%). No other genomic alteration reaches significance after accounting for multiple hypothesis testing, and no significant differences in tumor mutational burden are observed between HER2-low and HER2-0 tumors (median: 7.26 mutations/megabase vs. 7.60 mutations/megabase, p = 0.24). Here, we show that the genomic landscape of HER2-low and HER2-0 tumors does not differ significantly, apart from a higher ERBB2 copy count among HER2-low tumors, and a higher rate of ERBB2 hemideletions in HER2-0 tumors.
© 2023. The Author(s).
Conflict of interest statement
P.T. served as advisor/consultant for AstraZeneca, Daiichi-Sankyo, Gilead and Lilly. N.U.L. reports institutional research support from Genentech, Pfizer, Merck, Seattle Genetics, Zion Pharmaceuticals, Olema Pharmaceuticals, and AstraZeneca; consulting honoraria from Puma, Seattle Genetics, Daiichi-Sankyo, AstraZeneca, Denali Therapeutics, Prelude Therapeutics, Olema Pharmaceuticals, Aleta BioPharma, Affinia Therapeutics, Voyager Therapeutics, Janssen, Blueprint Medicines, Stemline/Menarini, and Artera Inc. and Reverie Labs;.; and royalties from UptoDate (book). S.M.T. reports consulting or advisory roles for Novartis, Pfizer, Merck, Eli Lilly, AstraZeneca, Genentech/Roche, Eisai, Sanofi, Bristol Myers Squibb, Seattle Genetics, CytomX Therapeutics, Daiichi-Sankyo, Gilead, Ellipses Pharma, 4D Pharma, OncoSec Medical Inc., BeyondSpring Pharmaceuticals, OncXerna, Zymeworks, Zentalis, Blueprint Medicines, Reveal Genomics, ARC Therapeutics, Infinity Therapeutics, Myovant, Zetagen, Umoja Biopharma, Artios Pharma, Menarini/Stemline, Aadi Biopharma, Bayer, Incyte Corp, and Jazz Pharmaceuticals; and research funding from Genentech/Roche, Merck, Exelixis, Pfizer, Lilly, Novartis, Bristol Myers Squibb, Eisai, AstraZeneca, Gilead, NanoString Technologies, Seattle Genetics, and OncoPep. G.C. reports honoraria for speaker’s engagement from Roche, Seattle Genetics, Novartis, Lilly, Pfizer, Foundation Medicine, NanoString, Samsung, Celltrion, BMS, MSD; honoraria for providing consultancy from Roche, Seattle Genetics, NanoString; honoraria for participating on the advisory boards of Roche, Lilly, Pfizer, Foundation Medicine, Samsung, Celltrion, Mylan; honoraria for writing engagement from Novartis and BMS; honoraria for participation in the Ellipsis Scientific Affairs Group; institutional research funding for conducting phase I and II clinical trials from Pfizer, Roche, Novartis, Sanofi, Celgene, Servier, Orion, AstraZeneca, Seattle Genetics, AbbVie, Tesaro, BMS, Merck Serono, Merck Sharp Dome, Janssen-Cilag, Philogen, Bayer, Medivation, and Medimmune. A.C.G.-C. reports research funding (to Institution) from AstraZeneca, Daiichi-Sankyo, Merck, Gilead Sciences, Zenith Epigenetics; and travel accommodations from Roche/Genentech. R.B.-S. reports receiving speaker bureau fees from Agilant, AstraZeneca, Daiichi-Sankyo, Eli Lilly, Pfizer, Novartis, Merck, and Roche. He has also served as a consultant/advisor for AstraZeneca, Eli Lilly, Libbs, Roche, Merck and has received support for attending medical conferences from AstraZeneca, Roche, Eli Lilly, Daiichi-Sankyo, and Merck. D.D. has served as a consultant for Novartis, on the Advisory Board for Oncology Analytics and receives research funding from Canon Inc. B.E.J. reports that he has served as a paid consultant to Novartis, Checkpoint Therapeutics, Hummingbird Diagnostics, Daiichi- Sankyo, AstraZeneca, G1 Therapeutics, BlueDotBio, GSK, Hengrui Therapeutics, Simcere Pharmaceutical, Jazz Pharmaceuticals, and Merus N.V. He is also an unpaid member of a Steering Committee for Pfizer. He receives research support from Cannon Medical Imaging. A.D.C. receives research support from Bayer AG. M.M. reports research support from Bayer AG and Janssen; consulting for Bayer, Delve, Interline, and Isabl; and royalties from Bayer and LabCorp. R.J. reports research support from Pfizer and Lilly and serves as an advisor to GE Health and Carrick Therapeutics. The remaining authors declare no competing interests.
Figures


References
-
- Gianni L, et al. Open-label, phase II, multicenter, randomized study of the efficacy and safety of two dose levels of pertuzumab, a human epidermal growth factor receptor 2 dimerization inhibitor, in patients with human epidermal growth factor receptor 2–negative metastatic breast cancer. J. Clin. Oncol. 2010;28:1131–1137. doi: 10.1200/JCO.2009.24.1661. - DOI - PMC - PubMed
-
- Tarantino, P. et al. Antibody-drug conjugates: smart chemotherapy delivery across tumor histologies. CA Cancer J. Clin.72, 165–182 (2022). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

