SARS-CoV-2 helicase might interfere with cellular nonsense-mediated RNA decay: insights from a bioinformatics study
- PMID: 37980504
- PMCID: PMC10657555
- DOI: 10.1186/s12863-023-01173-y
SARS-CoV-2 helicase might interfere with cellular nonsense-mediated RNA decay: insights from a bioinformatics study
Abstract
Background: Viruses employ diverse strategies to interfere with host defense mechanisms, including the production of proteins that mimic or resemble host proteins. This study aimed to analyze the similarities between SARS-CoV-2 and human proteins, investigate their impact on virus-host interactions, and elucidate underlying mechanisms.
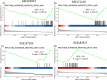
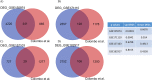
Results: Comparing the proteins of SARS-CoV-2 with human and mammalian proteins revealed sequence and structural similarities between viral helicase with human UPF1. The latter is a protein that is involved in nonsense-mediated RNA decay (NMD), an mRNA surveillance pathway which also acts as a cellular defense mechanism against viruses. Protein sequence similarities were also observed between viral nsp3 and human Poly ADP-ribose polymerase (PARP) family of proteins. Gene set enrichment analysis on transcriptomic data derived from SARS-CoV-2 positive samples illustrated the enrichment of genes belonging to the NMD pathway compared with control samples. Moreover, comparing transcriptomic data from SARS-CoV-2-infected samples with transcriptomic data derived from UPF1 knockdown cells demonstrated a significant overlap between datasets.
Conclusions: These findings suggest that helicase/UPF1 sequence and structural similarity might have the ability to interfere with the NMD pathway with pathogenic and immunological implications.
Keywords: Coronaviridae; Helicase; Nonsense-mediated RNA decay; RNA surveillance; SARS-CoV-2.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Modulation of UPF1 catalytic activity upon interaction of SARS-CoV-2 Nucleocapsid protein with factors involved in nonsense mediated-mRNA decay.Nucleic Acids Res. 2024 Nov 27;52(21):13325-13339. doi: 10.1093/nar/gkae829. Nucleic Acids Res. 2024. PMID: 39360627 Free PMC article.
-
Modulation of NBAS-Related Functions in the Early Response to SARS-CoV-2 Infection.Int J Mol Sci. 2023 Jan 30;24(3):2634. doi: 10.3390/ijms24032634. Int J Mol Sci. 2023. PMID: 36768954 Free PMC article.
-
Biochemical characterization of the RNA helicase UPF1 involved in nonsense-mediated mRNA decay.Methods Enzymol. 2012;511:255-74. doi: 10.1016/B978-0-12-396546-2.00012-7. Methods Enzymol. 2012. PMID: 22713324
-
Targeting of viral RNAs by Upf1-mediated RNA decay pathways.Curr Opin Virol. 2021 Apr;47:1-8. doi: 10.1016/j.coviro.2020.11.002. Epub 2020 Dec 17. Curr Opin Virol. 2021. PMID: 33341474 Free PMC article. Review.
-
Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay.Genes Cells. 2013 Mar;18(3):161-75. doi: 10.1111/gtc.12033. Epub 2013 Jan 28. Genes Cells. 2013. PMID: 23356578 Review.
Cited by
-
Inference of differential kinase interaction networks with KINference.Bioinformatics. 2025 Jul 1;41(7):btaf349. doi: 10.1093/bioinformatics/btaf349. Bioinformatics. 2025. PMID: 40579228 Free PMC article.
-
Unveiling the role of PUS7-mediated pseudouridylation in host protein interactions specific for the SARS-CoV-2 RNA genome.Mol Ther Nucleic Acids. 2023 Oct 25;34:102052. doi: 10.1016/j.omtn.2023.102052. eCollection 2023 Dec 12. Mol Ther Nucleic Acids. 2023. PMID: 38028201 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
