SERPINB3-MYC axis induces the basal-like/squamous subtype and enhances disease progression in pancreatic cancer
- PMID: 37980563
- PMCID: PMC10842852
- DOI: 10.1016/j.celrep.2023.113434
SERPINB3-MYC axis induces the basal-like/squamous subtype and enhances disease progression in pancreatic cancer
Abstract
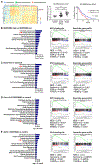
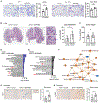
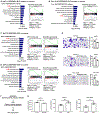
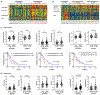
Pancreatic ductal adenocarcinoma (PDAC) exhibits distinct molecular subtypes: classical/progenitor and basal-like/squamous. Our study aimed to identify genes contributing to the development of the basal-like/squamous subtype, known for its aggressiveness. Transcriptome analyses revealed consistent upregulation of SERPINB3 in basal-like/squamous PDAC, correlating with reduced patient survival. SERPINB3 transgene expression in PDAC cells enhanced in vitro invasion and promoted lung metastasis in a mouse PDAC xenograft model. Metabolome analyses unveiled a metabolic signature linked to both SERPINB3 and the basal-like/squamous subtype, characterized by heightened carnitine/acylcarnitine and amino acid metabolism, associated with poor prognosis in patients with PDAC and elevated cellular invasiveness. Further analysis uncovered that SERPINB3 inhibited the cysteine protease calpain, a key enzyme in the MYC degradation pathway, and drove basal-like/squamous subtype and associated metabolic reprogramming through MYC activation. Our findings indicate that the SERPINB3-MYC axis induces the basal-like/squamous subtype, proposing SERPINB3 as a potential diagnostic and therapeutic target for this variant.
Keywords: BBOX1; CP: Cancer; CP: Metabolism; MYC; SERPINB3/SCCA1; amino acid; calpain; carnitine/acylcarnitine; metabolism; molecular subtype; pancreatic ductal adenocarcinoma; serine/cysteine proteinase inhibitor family B member 3.
Copyright © 2023. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interest.
Figures


References
-
- Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SGH, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung AH, et al. (2015). Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet 47, 1168–1178. 10.1038/ng.3398. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

