Ergolide mediates anti-cancer effects on metastatic uveal melanoma cells and modulates their cellular and extracellular vesicle proteomes
- PMID: 37981907
- PMCID: PMC10654492
- DOI: 10.12688/openreseurope.15973.2
Ergolide mediates anti-cancer effects on metastatic uveal melanoma cells and modulates their cellular and extracellular vesicle proteomes
Abstract
Background: Uveal melanoma is a poor prognosis cancer. Ergolide, a sesquiterpene lactone isolated from Inula Brittanica, exerts anti-cancer properties. The objective of this study was to 1) evaluate whether ergolide reduced metastatic uveal melanoma (MUM) cell survival/viability in vitro and in vivo; and 2) to understand the molecular mechanism of ergolide action.
Methods: Ergolide bioactivity was screened via long-term proliferation assay in UM/MUM cells and in zebrafish MUM xenograft models. Mass spectrometry profiled proteins modulated by ergolide within whole cell or extracellular vesicle (EVs) lysates of the OMM2.5 MUM cell line. Protein expression was analyzed by immunoblots and correlation analyses to UM patient survival used The Cancer Genome Atlas (TCGA) data.
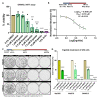
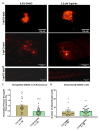
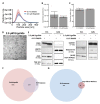
Results: Ergolide treatment resulted in significant, dose-dependent reductions (48.5 to 99.9%; p<0.0001) in OMM2.5 cell survival in vitro and of normalized primary zebrafish xenograft fluorescence (56%; p<0.0001) in vivo, compared to vehicle controls. Proteome-profiling of ergolide-treated OMM2.5 cells, identified 5023 proteins, with 52 and 55 proteins significantly altered at 4 and 24 hours, respectively ( p<0.05; fold-change >1.2). Immunoblotting of heme oxygenase 1 (HMOX1) and growth/differentiation factor 15 (GDF15) corroborated the proteomic data. Additional proteomics of EVs isolated from OMM2.5 cells treated with ergolide, detected 2931 proteins. There was a large overlap with EV proteins annotated within the Vesiclepedia compendium. Within the differentially expressed proteins, the proteasomal pathway was primarily altered. Interestingly, BRCA2 and CDKN1A Interacting Protein (BCCIP) and Chitinase Domain Containing 1 (CHID1), were the only proteins significantly differentially expressed by ergolide in both the OMM2.5 cellular and EV isolates and they displayed inverse differential expression in the cells versus the EVs.
Conclusions: Ergolide is a novel, promising anti-proliferative agent for UM/MUM. Proteomic profiling of OMM2.5 cellular/EV lysates identified candidate pathways elucidating the action of ergolide and putative biomarkers of UM, that require further examination.
Keywords: BRCA2 and CDKN1A Interacting Protein; Chitinase Domain Containing 1; Metastatic uveal melanoma; ergolide; extracellular vesicles.
Plain language summary
The most common form of adult eye cancer is uveal melanoma (UM). Once UM cancer cells spread to organs in the rest of the body, metastatic UM (MUM), there is a poor prognosis for patients with only one approved drug treatment. Hence, it is vital to better understand the cellular and extracellular proteins that regulate UM pathology in order to uncover biomarkers of disease and therapeutic targets. In this original study, we demonstrate a compound called ergolide is capable of severely reducing the metabolic activity and growth of UM cancer cells, grown as isolated monolayers. Ergolide was also able to reduce the growth of human MUM cells growing as tumors in transplanted zebrafish larvae. We identify that ergolide alters specific proteins found in the human UM cells. These proteins once analyzed in detail offer opportunities to understand how new treatment strategies can be developed for UM.
Copyright: © 2023 Sundaramurthi H et al.
Conflict of interest statement
No competing interests were disclosed.
Figures




References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
