ESR1 F404 Mutations and Acquired Resistance to Fulvestrant in ESR1-Mutant Breast Cancer
- PMID: 37982575
- PMCID: PMC10850945
- DOI: 10.1158/2159-8290.CD-22-1387
ESR1 F404 Mutations and Acquired Resistance to Fulvestrant in ESR1-Mutant Breast Cancer
Abstract
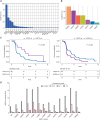
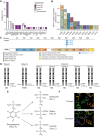
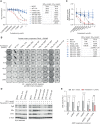
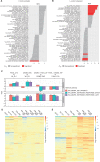
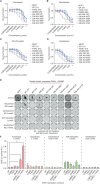
Fulvestrant is used to treat patients with hormone receptor-positive advanced breast cancer, but acquired resistance is poorly understood. PlasmaMATCH Cohort A (NCT03182634) investigated the activity of fulvestrant in patients with activating ESR1 mutations in circulating tumor DNA (ctDNA). Baseline ESR1 mutations Y537S are associated with poor outcomes and Y537C with good outcomes. Sequencing of baseline and EOT ctDNA samples (n = 69) revealed 3/69 (4%) patients acquired novel ESR1 F404 mutations (F404L, F404I, and F404V), in cis with activating mutations. In silico modeling revealed that ESR1 F404 contributes to fulvestrant binding to estrogen receptor-alpha (ERα) through a pi-stacking bond, with mutations disrupting this bond. In vitro analysis demonstrated that single F404L, E380Q, and D538G models were less sensitive to fulvestrant, whereas compound mutations D538G + F404L and E380Q + F404L were resistant. Several oral ERα degraders were active against compound mutant models. We have identified a resistance mechanism specific to fulvestrant that can be targeted by treatments in clinical development.
Significance: Novel F404 ESR1 mutations may be acquired to cause overt resistance to fulvestrant when combined with preexisting activating ESR1 mutations. Novel combinations of mutations in the ER ligand binding domain may cause drug-specific resistance, emphasizing the potential of similar drug-specific mutations to impact the efficacy of oral ER degraders in development. This article is featured in Selected Articles from This Issue, p. 201.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures

References
-
- Turner NC, Ro J, Andre F, Loi S, Verma S, Iwata H, et al. Palbociclib in hormone-receptor-positive advanced breast cancer. N Engl J Med 2015;373:209–19. - PubMed
-
- Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med 2019;380:1929–40. - PubMed
-
- Sledge GW Jr, Toi M, Neven P, Sohn J, Inoue K, Pivot X, et al. The effect of abemaciclib plus fulvestrant on overall survival in hormone receptor-positive, ERBB2-negative breast cancer that progressed on endocrine therapy-MONARCH 2: a randomized clinical trial. JAMA Oncol 2020;6:116–24. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

