Virulence attributes of successful methicillin-resistant Staphylococcus aureus lineages
- PMID: 37982596
- PMCID: PMC10732075
- DOI: 10.1128/cmr.00148-22
Virulence attributes of successful methicillin-resistant Staphylococcus aureus lineages
Abstract
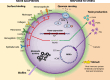
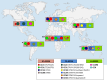
Methicillin-resistant Staphylococcus aureus (MRSA) is a leading cause of severe and often fatal infections. MRSA epidemics have occurred in waves, whereby a previously successful lineage has been replaced by a more fit and better adapted lineage. Selection pressures in both hospital and community settings are not uniform across the globe, which has resulted in geographically distinct epidemiology. This review focuses on the mechanisms that trigger the establishment and maintenance of current, dominant MRSA lineages across the globe. While the important role of antibiotic resistance will be mentioned throughout, factors which influence the capacity of S. aureus to colonize and cause disease within a host will be the primary focus of this review. We show that while MRSA possesses a diverse arsenal of toxins including alpha-toxin, the success of a lineage involves more than just producing toxins that damage the host. Success is often attributed to the acquisition or loss of genetic elements involved in colonization and niche adaptation such as the arginine catabolic mobile element, as well as the activity of regulatory systems, and shift metabolism accordingly (e.g., the accessory genome regulator, agr). Understanding exactly how specific MRSA clones cause prolonged epidemics may reveal targets for therapies, whereby both core (e.g., the alpha toxin) and acquired virulence factors (e.g., the Panton-Valentine leukocidin) may be nullified using anti-virulence strategies.
Keywords: gene regulation; metabolism; methicillin-resistant Staphylococcus aureus; mobile genetic elements; superantigens; toxins; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

