Mapping the configurational landscape and aggregation phase behavior of the tau protein fragment PHF6
- PMID: 37983502
- PMCID: PMC10691331
- DOI: 10.1073/pnas.2309995120
Mapping the configurational landscape and aggregation phase behavior of the tau protein fragment PHF6
Abstract
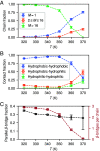
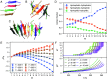
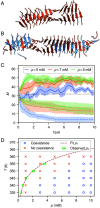
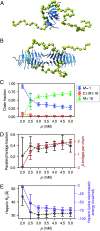
The PHF6 (Val-Gln-Ile-Val-Tyr-Lys) motif, found in all isoforms of the microtubule-associated protein tau, forms an integral part of ordered cores of amyloid fibrils formed in tauopathies and is thought to play a fundamental role in tau aggregation. Because PHF6 as an isolated hexapeptide assembles into ordered fibrils on its own, it is investigated as a minimal model for insight into the initial stages of aggregation of larger tau fragments. Even for this small peptide, however, the large length and time scales associated with fibrillization pose challenges for simulation studies of its dynamic assembly, equilibrium configurational landscape, and phase behavior. Here, we develop an accurate, bottom-up coarse-grained model of PHF6 for large-scale simulations of its aggregation, which we use to uncover molecular interactions and thermodynamic driving forces governing its assembly. The model, not trained on any explicit information about fibrillar structure, predicts coexistence of formed fibrils with monomers in solution, and we calculate a putative equilibrium phase diagram in concentration-temperature space. We also characterize the configurational and free energetic landscape of PHF6 oligomers. Importantly, we demonstrate with a model of heparin that this widely studied cofactor enhances the aggregation propensity of PHF6 by ordering monomers during nucleation and remaining associated with growing fibrils, consistent with experimentally characterized heparin-tau interactions. Overall, this effort provides detailed molecular insight into PHF6 aggregation thermodynamics and pathways and, furthermore, demonstrates the potential of modern multiscale modeling techniques to produce predictive models of amyloidogenic peptides simultaneously capturing sequence-specific effects and emergent aggregate structures.
Keywords: amyloid aggregation; multiscale modeling; tau protein.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
References
-
- Ballatore C., Lee V. M. Y., Trojanowski J. Q., Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat. Rev. Neurosci. 8, 663–672 (2007). - PubMed
-
- Goedert M., Eisenberg D. S., Crowther R. A., Propagation of tau aggregates and neurodegeneration. Annu. Rev. Neurosci. 40, 189–210 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

