Enhanced C-To-T and A-To-G Base Editing in Mitochondrial DNA with Engineered DdCBE and TALED
- PMID: 37984866
- PMCID: PMC10797475
- DOI: 10.1002/advs.202304113
Enhanced C-To-T and A-To-G Base Editing in Mitochondrial DNA with Engineered DdCBE and TALED
Abstract
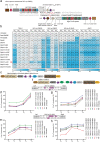
Mitochondrial base editing with DddA-derived cytosine base editor (DdCBE) is limited in the accessible target sequences and modest activity. Here, the optimized DdCBE tools is presented with improved editing activity and expanded C-to-T targeting scope by fusing DddA11 variant with different cytosine deaminases with single-strand DNA activity. Compared to previous DdCBE based on DddA11 variant alone, fusion of the activation-induced cytidine deaminase (AID) from Xenopus laevis not only permits cytosine editing of 5'-GC-3' sequence, but also elevates editing efficiency at 5'-TC-3', 5'-CC-3', and 5'-GC-3' targets by up to 25-, 10-, and 6-fold, respectively. Furthermore, the A-to-G editing efficiency is significantly improved by fusing the evolved DddA6 variant with TALE-linked deoxyadenosine deaminase (TALED). Notably, the authors introduce the reported high-fidelity mutations in DddA and add nuclear export signal (NES) sequences in DdCBE and TALED to reduce off-target editing in the nuclear and mitochondrial genome while improving on-target editing efficiency in mitochondrial DNA (mtDNA). Finally, these engineered mitochondrial base editors are shown to be efficient in installing mtDNA mutations in human cells or mouse embryos for disease modeling. Collectively, the study shows broad implications for the basic study and therapeutic applications of optimized DdCBE and TALED.
Keywords: DdCBE; TALED; disease modeling; mitochondrial base editing.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
