Fast satellite DNA evolution in Nothobranchius annual killifishes
- PMID: 37985497
- PMCID: PMC10661780
- DOI: 10.1007/s10577-023-09742-8
Fast satellite DNA evolution in Nothobranchius annual killifishes
Abstract
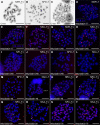
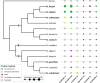
Satellite DNA (satDNA) is a rapidly evolving class of tandem repeats, with some monomers being involved in centromere organization and function. To identify repeats associated with (peri)centromeric regions, we investigated satDNA across Southern and Coastal clades of African annual killifishes of the genus Nothobranchius. Molecular cytogenetic and bioinformatic analyses revealed that two previously identified satellites, designated here as NkadSat01-77 and NfurSat01-348, are associated with (peri)centromeres only in one lineage of the Southern clade. NfurSat01-348 was, however, additionally detected outside centromeres in three members of the Coastal clade. We also identified a novel satDNA, NrubSat01-48, associated with (peri)centromeres in N. foerschi, N. guentheri, and N. rubripinnis. Our findings revealed fast turnover of satDNA associated with (peri)centromeres and different trends in their evolution in two clades of the genus Nothobranchius.
Keywords: Centromere drive; Constitutive heterochromatin; RepeatExplorer; Repetitive sequences; satDNA.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Sex chromosome differentiation via changes in the Y chromosome repeat landscape in African annual killifishes Nothobranchius furzeri and N. kadleci.Chromosome Res. 2022 Dec;30(4):309-333. doi: 10.1007/s10577-022-09707-3. Epub 2022 Oct 8. Chromosome Res. 2022. PMID: 36208359
-
Conserved satellite DNA motif and lack of interstitial telomeric sites in highly rearranged African Nothobranchius killifish karyotypes.J Fish Biol. 2023 Dec;103(6):1501-1514. doi: 10.1111/jfb.15550. Epub 2023 Sep 19. J Fish Biol. 2023. PMID: 37661806
-
Dissecting the Satellite DNA Landscape in Three Cactophilic Drosophila Sequenced Genomes.G3 (Bethesda). 2017 Aug 7;7(8):2831-2843. doi: 10.1534/g3.117.042093. G3 (Bethesda). 2017. PMID: 28659292 Free PMC article.
-
Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin.Gene. 2008 Feb 15;409(1-2):72-82. doi: 10.1016/j.gene.2007.11.013. Epub 2007 Dec 4. Gene. 2008. PMID: 18182173 Review.
-
Molecular Dynamics and Evolution of Centromeres in the Genus Equus.Int J Mol Sci. 2022 Apr 10;23(8):4183. doi: 10.3390/ijms23084183. Int J Mol Sci. 2022. PMID: 35457002 Free PMC article. Review.
Cited by
-
Multiple Origins of Sex Chromosomes in Nothobranchius Killifishes.Mol Ecol. 2025 Aug;34(16):e70029. doi: 10.1111/mec.70029. Epub 2025 Jul 24. Mol Ecol. 2025. PMID: 40702874 Free PMC article.
-
Independent evolution of satellite DNA sequences in homologous sex chromosomes of Neotropical armored catfish (Harttia).Commun Biol. 2025 Mar 30;8(1):524. doi: 10.1038/s42003-025-07891-6. Commun Biol. 2025. PMID: 40159539 Free PMC article.
-
Satellite DNA Mapping in Suliformes (Aves): Insights into the Evolution of the Multiple Sex Chromosome System in Sula spp.Genes (Basel). 2025 May 24;16(6):633. doi: 10.3390/genes16060633. Genes (Basel). 2025. PMID: 40565525 Free PMC article.
-
Repetitive DNAs and differentiation of the ZZ/ZW sex chromosome system in the combtail fish Belontia hasselti (Perciformes: Osphronemidae).BMC Ecol Evol. 2025 Mar 18;25(1):25. doi: 10.1186/s12862-025-02358-y. BMC Ecol Evol. 2025. PMID: 40098070 Free PMC article.
-
First karyotype description of Epiplatysspilargyreius (Duméril, 1861) with comments on chromosome evolution in the genus Epiplatys Gill, 1862 (Nothobranchiidae).Comp Cytogenet. 2025 Jun 19;19:109-115. doi: 10.3897/compcytogen.19.151345. eCollection 2025. Comp Cytogenet. 2025. PMID: 40636885 Free PMC article.
References
-
- Andrews S (2010) FastQC: a quality control tool for high throughput sequence data [Online]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 25 Sept 2019
-
- Ávila Robledillo L, Koblížková A, Novák P, Böttinger K, Vrbová I, Neumann P, Schubert I, Macas J. Satellite DNA in Vicia faba is characterized by remarkable diversity in its sequence composition, association with centromeres, and replication timing. Sci Rep. 2018;8:5838. doi: 10.1038/s41598-018-24196-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- 19-22346Y/Grantová Agentura České Republiky
- 19-22346Y/Grantová Agentura České Republiky
- 19-22346Y/Grantová Agentura České Republiky
- 19-22346Y/Grantová Agentura České Republiky
- 19-22346Y/Grantová Agentura České Republiky
- 19-22346Y/Grantová Agentura České Republiky
- 19-22346Y/Grantová Agentura České Republiky
- 19-22346Y/Grantová Agentura České Republiky
- 19-22346Y/Grantová Agentura České Republiky
- e-INFRA LM2018140/e-Infrastruktura CZ
- e-INFRA LM2018140/e-Infrastruktura CZ
- e-INFRA LM2018140/e-Infrastruktura CZ
- e-INFRA LM2018140/e-Infrastruktura CZ
- LM2018131/Projects of Large Research, Development and Innovations Infrastructures and the ELIXIR-CZ project
- LM2018131/Projects of Large Research, Development and Innovations Infrastructures and the ELIXIR-CZ project
- LM2018131/Projects of Large Research, Development and Innovations Infrastructures and the ELIXIR-CZ project
- LM2018131/Projects of Large Research, Development and Innovations Infrastructures and the ELIXIR-CZ project
- UJAR10MS/Spanish Ministry of Universities with European Union's NextGenerationEU funds
- 67985904/RVO of IAPG CAS
- 67985904/RVO of IAPG CAS
- 67985904/RVO of IAPG CAS
- 204069/Charles University Research Centre program
LinkOut - more resources
Full Text Sources

