Early-to-mid stage idiopathic Parkinson's disease shows enhanced cytotoxicity and differentiation in CD8 T-cells in females
- PMID: 37985656
- PMCID: PMC10662447
- DOI: 10.1038/s41467-023-43053-0
Early-to-mid stage idiopathic Parkinson's disease shows enhanced cytotoxicity and differentiation in CD8 T-cells in females
Abstract
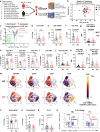
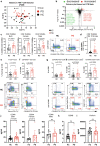
Neuroinflammation in the brain contributes to the pathogenesis of Parkinson's disease (PD), but the potential dysregulation of peripheral immunity has not been systematically investigated for idiopathic PD (iPD). Here we showed an elevated peripheral cytotoxic immune milieu, with more terminally-differentiated effector memory (TEMRA) CD8 T, CD8+ NKT cells and circulating cytotoxic molecules in fresh blood of patients with early-to-mid iPD, especially females, after analyzing > 700 innate and adaptive immune features. This profile, also reflected by fewer CD8+FOXP3+ T cells, was confirmed in another subcohort. Co-expression between cytotoxic molecules was selectively enhanced in CD8 TEMRA and effector memory (TEM) cells. Single-cell RNA-sequencing analysis demonstrated the accelerated differentiation within CD8 compartments, enhanced cytotoxic pathways in CD8 TEMRA and TEM cells, while CD8 central memory (TCM) and naïve cells were already more-active and transcriptionally-reprogrammed. Our work provides a comprehensive map of dysregulated peripheral immunity in iPD, proposing candidates for early diagnosis and treatments.
© 2023. The Author(s).
Conflict of interest statement
Pending patent application on the protection of biomarkers for Parkinson’s disease (patent applicant: Luxembourg Institute of Health; inventors: F.Q.H., M.O. and R.K.; EP Patent Application No. 23203381.1 entitled “Early biomarker for Parkinson’s disease”). The remaining authors of this work declare no competing interests.
Figures





References
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
- CORE/14/BM/8231540/GeDES/Fonds National de la Recherche Luxembourg (National Research Fund)
- Tregbar/11228353/Fonds National de la Recherche Luxembourg (National Research Fund)
- PRIPRIDDE/11012546/NEXTIMMUNE/Fonds National de la Recherche Luxembourg (National Research Fund)
- PRIDE/10907093/CRITICS/Fonds National de la Recherche Luxembourg (National Research Fund)
- PRIDE/14254520/i2TRON/Fonds National de la Recherche Luxembourg (National Research Fund)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

