Genomic data resources of the Brain Somatic Mosaicism Network for neuropsychiatric diseases
- PMID: 37985666
- PMCID: PMC10662356
- DOI: 10.1038/s41597-023-02645-7
Genomic data resources of the Brain Somatic Mosaicism Network for neuropsychiatric diseases
Abstract
Somatic mosaicism is defined as an occurrence of two or more populations of cells having genomic sequences differing at given loci in an individual who is derived from a single zygote. It is a characteristic of multicellular organisms that plays a crucial role in normal development and disease. To study the nature and extent of somatic mosaicism in autism spectrum disorder, bipolar disorder, focal cortical dysplasia, schizophrenia, and Tourette syndrome, a multi-institutional consortium called the Brain Somatic Mosaicism Network (BSMN) was formed through the National Institute of Mental Health (NIMH). In addition to genomic data of affected and neurotypical brains, the BSMN also developed and validated a best practices somatic single nucleotide variant calling workflow through the analysis of reference brain tissue. These resources, which include >400 terabytes of data from 1087 subjects, are now available to the research community via the NIMH Data Archive (NDA) and are described here.
© 2023. The Author(s).
Conflict of interest statement
JVM is an inventor on patent US6150160, is a paid consultant for Gilead Sciences, and serves on the scientific advisory board of Tessera Therapeutics Inc., where he is paid as a consultant and has equity options. He has also recently served on the American Society of Human Genetics Board of Directors. CAW is a paid consultant to Third Rock Ventures and Flagship Pioneering, and is on the Clinical Advisory Board of Maze Therapeutics. DRW is on the Scientific Advisory Boards of Sage Therapeutics and Pasithea Therapeutics. The other authors do not declare competing interests. This article was prepared while MAP was employed at Sage Bionetworks. The opinions expressed in this article are the author’s own and do not reflect the view of the National Institute on Aging, the National Institutes of Health, the Department of Health and Human Services, or the United States government.
Figures
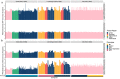
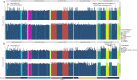
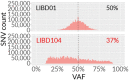
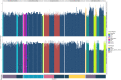
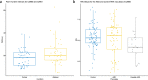
References
Publication types
MeSH terms
Grants and funding
- U01 MH106883/MH/NIMH NIH HHS/United States
- R01 MH124890/MH/NIMH NIH HHS/United States
- T32 GM007445/GM/NIGMS NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- K99 HD111686/HD/NICHD NIH HHS/United States
- U01 MH106884/MH/NIMH NIH HHS/United States
- T32 GM007544/GM/NIGMS NIH HHS/United States
- U01 MH106874/MH/NIMH NIH HHS/United States
- U01 MH106892/MH/NIMH NIH HHS/United States
- U01 MH108898/MH/NIMH NIH HHS/United States
- T32 HG002295/HG/NHGRI NIH HHS/United States
- U01 MH106891/MH/NIMH NIH HHS/United States
- R21 MH134401/MH/NIMH NIH HHS/United States
- U01 MH106876/MH/NIMH NIH HHS/United States
- U01 MH106882/MH/NIMH NIH HHS/United States
- U01 MH106893/MH/NIMH NIH HHS/United States

