Megakaryocyte- and erythroblast-specific cell-free DNA patterns in plasma and platelets reflect thrombopoiesis and erythropoiesis levels
- PMID: 37985773
- PMCID: PMC10662131
- DOI: 10.1038/s41467-023-43310-2
Megakaryocyte- and erythroblast-specific cell-free DNA patterns in plasma and platelets reflect thrombopoiesis and erythropoiesis levels
Abstract
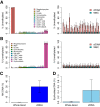
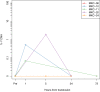
Circulating cell-free DNA (cfDNA) fragments are a biological analyte with extensive utility in diagnostic medicine. Understanding the source of cfDNA and mechanisms of release is crucial for designing and interpreting cfDNA-based liquid biopsy assays. Using cell type-specific methylation markers as well as genome-wide methylation analysis, we determine that megakaryocytes, the precursors of anuclear platelets, are major contributors to cfDNA (~26%), while erythroblasts contribute 1-4% of cfDNA in healthy individuals. Surprisingly, we discover that platelets contain genomic DNA fragments originating in megakaryocytes, contrary to the general understanding that platelets lack genomic DNA. Megakaryocyte-derived cfDNA is increased in pathologies involving increased platelet production (Essential Thrombocythemia, Idiopathic Thrombocytopenic Purpura) and decreased upon reduced platelet production due to chemotherapy-induced bone marrow suppression. Similarly, erythroblast cfDNA is reflective of erythrocyte production and is elevated in patients with thalassemia. Megakaryocyte- and erythroblast-specific DNA methylation patterns can thus serve as biomarkers for pathologies involving increased or decreased thrombopoiesis and erythropoiesis, which can aid in determining the etiology of aberrant levels of erythrocytes and platelets.
© 2023. The Author(s).
Conflict of interest statement
J.M., R.S., B.G., and Y.D. are inventors of patents describing methylation markers and their use for cfDNA analysis. All remaining authors have declared no conflicts of interest.
Figures



References
-
- Lo, Y. M. D. et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 10.1126/scitranslmed.3001720 (2010). - PubMed
-
- Dor, Y. & Cedar, H. Principles of DNA methylation and their implications for biology and medicine. Lancet10.1016/S0140-6736(18)31268-6 (2018). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

