How AlphaFold2 shaped the structural coverage of the human transmembrane proteome
- PMID: 37985809
- PMCID: PMC10662385
- DOI: 10.1038/s41598-023-47204-7
How AlphaFold2 shaped the structural coverage of the human transmembrane proteome
Abstract
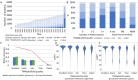
AlphaFold2 (AF2) provides a 3D structure for every known or predicted protein, opening up new prospects for virtually every field in structural biology. However, working with transmembrane protein molecules pose a notorious challenge for scientists, resulting in a limited number of experimentally determined structures. Consequently, algorithms trained on this finite training set also face difficulties. To address this issue, we recently launched the TmAlphaFold database, where predicted AlphaFold2 structures are embedded into the membrane plane and a quality assessment (plausibility of the membrane-embedded structure) is provided for each prediction using geometrical evaluation. In this paper, we analyze how AF2 has improved the structural coverage of membrane proteins compared to earlier years when only experimental structures were available, and high-throughput structure prediction was greatly limited. We also evaluate how AF2 can be used to search for (distant) homologs in highly diverse protein families. By combining quality assessment and homology search, we can pinpoint protein families where AF2 accuracy is still limited, and experimental structure determination would be desirable.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

