DNMT3a-mediated methylation of PPARγ promote intervertebral disc degeneration by regulating the NF-κB pathway
- PMID: 37986543
- PMCID: PMC10826446
- DOI: 10.1111/jcmm.18048
DNMT3a-mediated methylation of PPARγ promote intervertebral disc degeneration by regulating the NF-κB pathway
Abstract
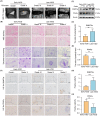
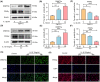
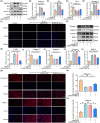
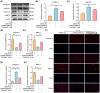
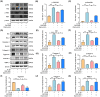
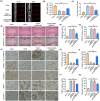
Intervertebral disc degeneration (IVDD) is a common chronic musculoskeletal disease that causes chronic low back pain and imposes an immense financial strain on patients. The pathological mechanisms underlying IVDD have not been fully elucidated. The development of IVDD is closely associated with abnormal epigenetic changes, suggesting that IVDD progression may be controlled by epigenetic mechanisms. Consequently, this study aimed to investigate the role of epigenetic regulation, including DNA methyltransferase 3a (DNMT3a)-mediated methylation and peroxisome proliferator-activated receptor γ (PPARγ) inhibition, in IVDD development. The expression of DNMT3a and PPARγ in early and late IVDD of nucleus pulposus (NP) tissues was detected using immunohistochemistry and western blotting analyses. Cellularly, DNMT3a inhibition significantly inhibited IL-1β-induced apoptosis and extracellular matrix (ECM) degradation in rat NP cells. Pretreatment with T0070907, a specific inhibitor of PPARγ, significantly reversed the anti-apoptotic and ECM degradation effects of DNMT3a inhibition. Mechanistically, DNMT3a modified PPARγ promoter hypermethylation to activate the nuclear factor-κB (NF-κB) pathway. DNMT3a inhibition alleviated IVDD progression. Conclusively, the results of this study show that DNMT3a activates the NF-κB pathway by modifying PPARγ promoter hypermethylation to promote apoptosis and ECM degradation. Therefore, we believe that the ability of DNMT3a to mediate the PPARγ/NF-κB axis may provide new ideas for the potential pathogenesis of IVDD and may become an attractive target for the treatment of IVDD.
Keywords: DNMT3a; PPARγ; intervertebral disc degeneration; low back pain.
© 2023 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that there are no conficts of interest.
Figures



Similar articles
-
Ginsenoside Rg1 relieves rat intervertebral disc degeneration and inhibits IL-1β-induced nucleus pulposus cell apoptosis and inflammation via NF-κB signaling pathway.In Vitro Cell Dev Biol Anim. 2024 Mar;60(3):287-299. doi: 10.1007/s11626-024-00883-6. Epub 2024 Mar 14. In Vitro Cell Dev Biol Anim. 2024. PMID: 38485818 Free PMC article.
-
5-Azacytidine inhibits endoplasmic reticulum stress and apoptosis of nucleus pulposus cells by preserving PPARγ via promoter demethylation.In Vitro Cell Dev Biol Anim. 2025 Mar;61(3):288-297. doi: 10.1007/s11626-025-01021-6. Epub 2025 Mar 18. In Vitro Cell Dev Biol Anim. 2025. PMID: 40102314
-
Isoliquiritigenin mitigates intervertebral disc degeneration induced by oxidative stress and mitochondrial impairment through a PPARγ-dependent pathway.Free Radic Biol Med. 2024 Nov 20;225:98-111. doi: 10.1016/j.freeradbiomed.2024.10.001. Epub 2024 Oct 2. Free Radic Biol Med. 2024. PMID: 39366471
-
NF-κB signalling pathways in nucleus pulposus cell function and intervertebral disc degeneration.Cell Prolif. 2021 Jul;54(7):e13057. doi: 10.1111/cpr.13057. Epub 2021 May 24. Cell Prolif. 2021. PMID: 34028920 Free PMC article. Review.
-
Roles of p38 MAPK signalling in intervertebral disc degeneration.Cell Prolif. 2023 Aug;56(8):e13438. doi: 10.1111/cpr.13438. Epub 2023 Mar 5. Cell Prolif. 2023. PMID: 36872558 Free PMC article. Review.
Cited by
-
LRP1 mitigates intervertebral disc degeneration by inhibiting endoplasmic reticulum stress through stabilizing the PPARγ.J Orthop Translat. 2025 Jan 16;50:196-210. doi: 10.1016/j.jot.2024.12.009. eCollection 2025 Jan. J Orthop Translat. 2025. PMID: 39895867 Free PMC article.
-
Nucleotide polymorphism-based study utilizes human plasma liposomes to discover potential therapeutic targets for intervertebral disc disease.Front Endocrinol (Lausanne). 2024 Aug 15;15:1403523. doi: 10.3389/fendo.2024.1403523. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39211445 Free PMC article.
-
PPARs in Clinical Experimental Medicine after 35 Years of Worldwide Scientific Investigations and Medical Experiments.Biomolecules. 2024 Jul 1;14(7):786. doi: 10.3390/biom14070786. Biomolecules. 2024. PMID: 39062500 Free PMC article. Review.
-
Mulberroside A mitigates intervertebral disc degeneration by inhibiting MAPK and modulating Ppar-γ/NF-κB pathways.J Inflamm (Lond). 2024 Aug 28;21(1):32. doi: 10.1186/s12950-024-00398-7. J Inflamm (Lond). 2024. PMID: 39198816 Free PMC article.
References
-
- Guo Q, Zhu D, Wang Y, et al. Targeting STING attenuates ROS induced intervertebral disc degeneration. Osteoarthr Cartil. 2021;29(8):1213‐1224. - PubMed
-
- Binch ALA, Fitzgerald JC, Growney EA, Barry F. Cell‐based strategies for IVD repair: clinical progress and translational obstacles. Nat Rev Rheumatol. 2021;17(3):158‐175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

