This is a preprint.
2D and 3D multiplexed subcellular profiling of nuclear instability in human cancer
- PMID: 37986801
- PMCID: PMC10659270
- DOI: 10.1101/2023.11.07.566063
2D and 3D multiplexed subcellular profiling of nuclear instability in human cancer
Abstract
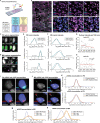
Nuclear atypia, including altered nuclear size, contour, and chromatin organization, is ubiquitous in cancer cells. Atypical primary nuclei and micronuclei can rupture during interphase; however, the frequency, causes, and consequences of nuclear rupture are unknown in most cancers. We demonstrate that nuclear envelope rupture is surprisingly common in many human cancers, particularly glioblastoma. Using highly-multiplexed 2D and super-resolution 3D-imaging of glioblastoma tissues and patient-derived xenografts and cells, we link primary nuclear rupture with reduced lamin A/C and micronuclear rupture with reduced lamin B1. Moreover, ruptured glioblastoma cells activate cGAS-STING-signaling involved in innate immunity. We observe that local patterning of cell states influences tumor spatial organization and is linked to both lamin expression and rupture frequency, with neural-progenitor-cell-like states exhibiting the lowest lamin A/C levels and greatest susceptibility to primary nuclear rupture. Our study reveals that nuclear instability is a core feature of cancer, and links nuclear integrity, cell state, and immune signaling.
Conflict of interest statement
Declaration of Interests PKS is a co-founder and member of the BOD of Glencoe Software, member of the BOD for Applied Biomath, and member of the SAB for RareCyte, NanoString, and Montai Health; he holds equity in Glencoe, Applied Biomath, and RareCyte. PKS consults for Merck and the Sorger Laboratory has received research funding from Novartis and Merck in the past five years. The DFCI receives funding for KLL’s research from the following entities: BMS, Lilly. KLL is co-founder of Travera Inc. KLL receives consulting fees from BMS, Travera, Integragen, Blaze Biosciences and BMS. DFCI and KLL have patents related to molecular diagnostics of cancer. D.P. is a member of the Volastra Therapeutics scientific advisory board. The other authors declare no outside interests.
Figures






References
-
- Zink D., Fischer A. H. & Nickerson J. A. Nuclear structure in cancer cells. Nat Rev Cancer 4, 677–687 (2004). - PubMed
-
- Fletcher C. D. M. Diagnostic Histopathology of Tumors. (Elsevier, 2013).
-
- Müller J. Ueber den feinern Bau und die Formen der krankhaften Geschwülste. (G. Reimer, 1838).
-
- Kato H. & Sandberg A. A. Chromosome pulverization in human cells with micronuclei. J Natl Cancer Inst 40, 165–179 (1968). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
