This is a preprint.
Transcriptomics and chromatin accessibility in multiple African population samples
- PMID: 37986808
- PMCID: PMC10659267
- DOI: 10.1101/2023.11.04.564839
Transcriptomics and chromatin accessibility in multiple African population samples
Abstract
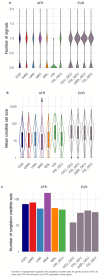
Mapping the functional human genome and impact of genetic variants is often limited to European-descendent population samples. To aid in overcoming this limitation, we measured gene expression using RNA sequencing in lymphoblastoid cell lines (LCLs) from 599 individuals from six African populations to identify novel transcripts including those not represented in the hg38 reference genome. We used whole genomes from the 1000 Genomes Project and 164 Maasai individuals to identify 8,881 expression and 6,949 splicing quantitative trait loci (eQTLs/sQTLs), and 2,611 structural variants associated with gene expression (SV-eQTLs). We further profiled chromatin accessibility using ATAC-Seq in a subset of 100 representative individuals, to identity chromatin accessibility quantitative trait loci (caQTLs) and allele-specific chromatin accessibility, and provide predictions for the functional effect of 78.9 million variants on chromatin accessibility. Using this map of eQTLs and caQTLs we fine-mapped GWAS signals for a range of complex diseases. Combined, this work expands global functional genomic data to identify novel transcripts, functional elements and variants, understand population genetic history of molecular quantitative trait loci, and further resolve the genetic basis of multiple human traits and disease.
Conflict of interest statement
Competing Interests PF is a member of the scientific advisory boards of Fabric Genomics, Inc., and Eagle Genomics, Ltd. AK is on the scientific advisory board of PatchBio, SerImmune, AINovo, TensorBio and OpenTargets, was a paid consultant with Illumina and owns shares in DeepGenomics, Immunai, Illumina, PatchBio and Freenome. SBM is a paid consultant for BioMarin, Tenaya Therapeutics and MyOme.
Figures









References
-
- Genetic effects on gene expression across human tissues | Nature. https://www.nature.com/articles/nature24277.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
