This is a preprint.
Are Escherichia coli causing recurrent cystitis just ordinary Uropathogenic E. coli (UPEC) strains?
- PMID: 37986820
- PMCID: PMC10659292
- DOI: 10.1101/2023.11.08.566351
Are Escherichia coli causing recurrent cystitis just ordinary Uropathogenic E. coli (UPEC) strains?
Update in
-
Are Escherichia coli causing recurrent cystitis just ordinary uropathogenic E. coli (UPEC) strains?Virulence. 2025 Dec;16(1):2444689. doi: 10.1080/21505594.2024.2444689. Epub 2024 Dec 26. Virulence. 2025. PMID: 39726097 Free PMC article.
Abstract
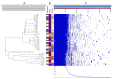
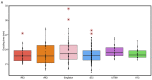
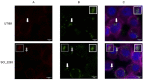
Specific determinants associated with Uropathogenic Escherichia coli (UPEC) causing recurrent cystitis are still poorly characterized. The aims of this study were (i) to describe genomic and phenotypic traits associated with recurrence using a large collection of recurrent and paired sporadic UPEC isolates, and (ii) to explore within-host genomic adaptation associated with recurrence using series of 2 to 5 sequential UPEC isolates. Whole genome comparative analyses between 24 recurrent cystitis isolates (RCIs) and 24 phylogenetically paired sporadic cystitis isolates (SCIs) suggested a lower prevalence of putative mobile genetic elements (MGE) in RCIs, such as plasmids and prophages. The intra-patient evolution of the 24 RCI series over time was characterized by SNP occurrence in genes involved in metabolism or membrane transport, and by plasmid loss in 5 out of the 24 RCI series. Genomic evolution occurred early in the course of recurrence, suggesting rapid adaptation to strong selection pressure in the urinary tract. However, RCIs did not exhibit specific virulence factor determinants and could not be distinguished from SCIs by their fitness, biofilm formation, or ability to invade HTB-9 bladder epithelial cells. Taken together, these results suggest a rapid but not convergent adaptation of RCIs that involves both strain- and host-specific characteristics.
Figures









References
-
- Ejrnaes K, Sandvang D, Lundgren B, Ferry S, Holm S, Monsen T, et al. Pulsed-Field Gel Electrophoresis Typing of Escherichia coli Strains from Samples Collected before and after Pivmecillinam or Placebo Treatment of Uncomplicated Community-Acquired Urinary Tract Infection in Women. J Clin Microbiol. 2006;44: 1776–1781. doi:10.1128/JCM.44.5.1776-1781.2006 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
