This is a preprint.
MSGene: Derivation and validation of a multistate model for lifetime risk of coronary artery disease using genetic risk and the electronic health record
- PMID: 37986972
- PMCID: PMC10659503
- DOI: 10.1101/2023.11.08.23298229
MSGene: Derivation and validation of a multistate model for lifetime risk of coronary artery disease using genetic risk and the electronic health record
Update in
-
MSGene: a multistate model using genetic risk and the electronic health record applied to lifetime risk of coronary artery disease.Nat Commun. 2024 Jun 7;15(1):4884. doi: 10.1038/s41467-024-49296-9. Nat Commun. 2024. PMID: 38849421 Free PMC article.
Abstract
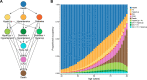
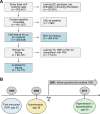
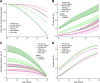
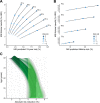
Currently, coronary artery disease (CAD) is the leading cause of death among adults worldwide. Accurate risk stratification can support optimal lifetime prevention. We designed a novel and general multistate model (MSGene) to estimate age-specific transitions across 10 cardiometabolic states, dependent on clinical covariates and a CAD polygenic risk score. MSGene supports decision making about CAD prevention related to any of these states. We analyzed longitudinal data from 480,638 UK Biobank participants and compared predicted lifetime risk with the 30-year Framingham risk score. MSGene improved discrimination (C-index 0.71 vs 0.66), age of high-risk detection (C-index 0.73 vs 0.52), and overall prediction (RMSE 1.1% vs 10.9%), with external validation. We also used MSGene to refine estimates of lifetime absolute risk reduction from statin initiation. Our findings underscore the potential public health value of our novel multistate model for accurate lifetime CAD risk estimation using clinical factors and increasingly available genetics.
Conflict of interest statement
DISCLOSURES During the course of the project, M.W.Y. became a full-time employee of GSK. A.C.F. is co-founder of Goodpath. PTE reports personal fees from Bayer AG, Novartis, and MyoKardia. GP holds equity in Phaeno Biotechnologies, is on the SAB of RealmIDX and currently consults for Delphi Diagnostics. P.N. reports research grants from Allelica, Apple, Amgen,Boston Scientific, Genentech / Roche, and Novartis, personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Foresite Labs, Genentech / Roche, GV, HeartFlow, Magnet Biomedicine, and Novartis, scientific advisory board membership of Esperion Therapeutics, Preciseli, and TenSixteen Bio, scientific co-founder of TenSixteen Bio, equity in MyOme, Preciseli, and TenSixteen Bio, and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. The remaining authors have nothing to disclose.
Figures

References
-
- Tsao CW, Aday AW, Almarzooq ZI, Anderson CAM, Arora P, Avery CL, Baker-Smith CM, Beaton AZ, Boehme AK, Buxton AE, Commodore-Mensah Y, Elkind MSV, Evenson KR, Eze-Nliam C, Fugar S, Generoso G, Heard DG, Hiremath S, Ho JE, Kalani R, Kazi DS, Ko D, Levine DA, Liu J, Ma J, Magnani JW, Michos ED, Mussolino ME, Navaneethan SD, Parikh NI, Poudel R, Rezk-Hanna M, Roth GA, Shah NS, St-Onge M-P, Thacker EL, Virani SS, Voeks JH, Wang N-Y, Wong ND, Wong SS, Yaffe K, Martin SS, Subcommittee on behalf of the AHAC on E and PSC and SS. Heart Disease and Stroke Statistics—2023 Update: A Report From the American Heart Association. Circulation [Internet]. 2023. [cited 2023 May 20];Available from: 10.1161/CIR.0000000000001123 - DOI - PubMed
-
- Lloyd-Jones DM, Leip EP, Larson MG, D’Agostino RB, Beiser A, Wilson PWF, Wolf PA, Levy D. Prediction of Lifetime Risk for Cardiovascular Disease by Risk Factor Burden at 50 Years of Age. Circulation. 2006;113:791–798. - PubMed
-
- Bundy JD, Ning H, Zhong VW, Paluch AE, Lloyd-Jones DM, Wilkins JT, Allen NB. Cardiovascular Health Score and Lifetime Risk of Cardiovascular Disease. Circulation: Cardiovascular Quality and Outcomes [Internet]. 2020. [cited 2023 Jun 13];Available from: 10.1161/CIRCOUTCOMES.119.006450 - DOI - PMC - PubMed
-
- Grundy SM, Stone NJ, Bailey AL, Beam C, Birtcher KK, Blumenthal RS, Braun LT, de Ferranti S, Faiella-Tommasino J, Forman DE, Goldberg R, Heidenreich PA, Hlatky MA, Jones DW, Lloyd-Jones D, Lopez-Pajares N, Ndumele CE, Orringer CE, Peralta CA, Saseen JJ, Smith SC, Sperling L, Virani SS, Yeboah J. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/ APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: Executive Summary. Circulation. 2019;139:e1082–e1143. - PMC - PubMed
Publication types
Grants and funding
- R01 HL139731/HL/NHLBI NIH HHS/United States
- R21 HG010748/HG/NHGRI NIH HHS/United States
- T32 HG010464/HG/NHGRI NIH HHS/United States
- R01 HL164629/HL/NHLBI NIH HHS/United States
- R01 HL157635/HL/NHLBI NIH HHS/United States
- R01 HL148050/HL/NHLBI NIH HHS/United States
- R01 HL092577/HL/NHLBI NIH HHS/United States
- R01 CA244569/CA/NCI NIH HHS/United States
- R01 CA227237/CA/NCI NIH HHS/United States
- R01 HL148565/HL/NHLBI NIH HHS/United States
- U01 HG011719/HG/NHGRI NIH HHS/United States
- K08 HL161448/HL/NHLBI NIH HHS/United States
- K23 HL169839/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous
