Microautophagy regulated by STK38 and GABARAPs is essential to repair lysosomes and prevent aging
- PMID: 37987447
- PMCID: PMC10702834
- DOI: 10.15252/embr.202357300
Microautophagy regulated by STK38 and GABARAPs is essential to repair lysosomes and prevent aging
Abstract
Lysosomes are degradative organelles and signaling hubs that maintain cell and tissue homeostasis, and lysosomal dysfunction is implicated in aging and reduced longevity. Lysosomes are frequently damaged, but their repair mechanisms remain unclear. Here, we demonstrate that damaged lysosomal membranes are repaired by microautophagy (a process termed "microlysophagy") and identify key regulators of the first and last steps. We reveal the AGC kinase STK38 as a novel microlysophagy regulator. Through phosphorylation of the scaffold protein DOK1, STK38 is specifically required for the lysosomal recruitment of the AAA+ ATPase VPS4, which terminates microlysophagy by promoting the disassembly of ESCRT components. By contrast, microlysophagy initiation involves non-canonical lipidation of ATG8s, especially the GABARAP subfamily, which is required for ESCRT assembly through interaction with ALIX. Depletion of STK38 and GABARAPs accelerates DNA damage-induced cellular senescence in human cells and curtails lifespan in C. elegans, respectively. Thus, microlysophagy is regulated by STK38 and GABARAPs and could be essential for maintaining lysosomal integrity and preventing aging.
Keywords: ESCRT; lysosome; microautophagy; non-canonical ATG8 lipidation.
© 2023 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
TY and SN are the founders of AutoPhagyGO.
Figures
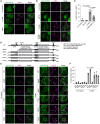
Representative images of transiently expressed mNeonGreen (mNG)‐STK38 (green) and immunostained LAMP1 (magenta) in HeLa cells treated with LLOMe (1 mM for 1 h, then incubated for 3 h after washout) or EBSS (for 4 h). Scale bars: 10 μm.
Representative images of stably expressed mNG‐STK38 (green) and immunostained LAMP1 (magenta) in HeLa cells treated with LLOMe (1 mM for 1 h) with or without pre‐treatment with BAPTA‐AM (10 μM for 1 h). Scale bars: 10 μm.
Quantification of colocalization between mNG‐STK38 and LAMP1 shown in (B). ≥ 50 cells were analyzed per experiment for each condition.
Schematic diagram of WT and mutant STK38.
Representative images of transiently expressed WT or mutant mNG‐STK38 (green) and immunostained LAMP1 (magenta) in HeLa cells treated with LLOMe (1 mM for 1 h, then incubated for 3 h after washout). Scale bars: 10 μm.
Quantification of colocalization between mNG‐STK38 and LAMP1 shown in (E). ≥ 50 cells were analyzed per experiment for each condition.
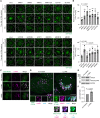
Representative images of stably expressed GFP‐Gal3 (green) in HeLa cells transfected with siRNAs of Hippo pathway components. Cells were treated with LLOMe (1 mM for 1 h, then incubated for the indicated number of hours after washout). Scale bars: 10 μm.
Percentage of residual GFP‐Gal3 dots (10 h/0 h [%]) in (A). ≥ 100 cells were analyzed per experiment for each condition.
Representative images of transiently expressed mNG‐MOB2 (green) and immunostained LAMP1 (magenta) in HeLa cells treated with LLOMe (1 mM for 1 h, then incubated for 3 h after washout) or EBSS (for 4 h). Scale bars: 10 μm.
Representative images of transiently expressed mNG‐STK38 (green), immunostained LAMP1 (magenta) and Gal3 (cyan) in HeLa cells treated with LLOMe (1 mM for 1 h, then incubated for 3 h after washout). Scale bars: 10 μm.
Representative immunoblot of phosphorylated STK38 T444 (pT444) in HeLa cells. Cells were treated with LLOMe (1 mM for 30 min). Asterisks in STK38 and STK38 pT444 blots represent STK38L and STK38L pT442, respectively.
Quantification of phosphorylation of STK38 T444 shown in (E).
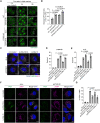
Representative images of stably expressed GFP‐Gal3 (green) in siSTK38‐transfected WT or FIP200 KO HeLa cells. Cells were treated with LLOMe (1 mM for 1 h, then incubated for the indicated number of hours after washout). Scale bars: 10 μm.
Percentage of residual GFP‐Gal3 dots (10 h/0 h [%]) in (A). ≥ 80 cells were analyzed per experiment for each condition.
Representative images of immunostained CHMP4B (green), ALIX (magenta), and DAPI (blue) in siSTK38‐transfected HeLa cells. Cells were treated with LLOMe (1 mM for 30 min or 1 h). Scale bars: 10 μm.
Quantification of CHMP4B dots shown in (C). ≥ 100 cells were analyzed per experiment for each condition.
Quantification of ALIX dots shown in (C). ≥ 100 cells were analyzed per experiment for each condition.
Representative images of immunostained VPS4 (green), LAMP1 (magenta), and DAPI (blue) in siSTK38‐transfected HeLa cells. Cells were treated with LLOMe (1 mM for 30 min or 1 h). Scale bars: 10 μm.
Quantification of co‐localization between VPS4 and LAMP1 shown in (F). ≥ 50 cells were analyzed per experiment for each condition.
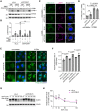
Representative immunoblots of LC3B in siSTK38‐transfected U2OS cells. Cells were incubated in normal growth media (GM) or EBSS with or without bafilomycinA1 (BafA1) for 2 h.
Quantification of LC3B shown in (A).
Representative images of stably expressed mNG‐ULK1 (green), LAMP1 (magenta), and DAPI (blue) in siSTK38‐transfected HeLa cells. Cells were treated with LLOMe (1 mM for 1 h, then incubated for 3 h after washout). Scale bars: 10 μm.
Quantification of co‐localization between mNG‐ULK1 and LAMP1 shown in (C). ≥ 50 cells were analyzed per experiment for each condition.
Representative images of stably expressed TFEB‐mNG (green) and DAPI (blue) in siSTK38‐transfected HeLa cells. Cells were treated with LLOMe (1 mM for 1 h, then incubated for 3 h after washout). Scale bars: 10 μm.
The nucleus/cytoplasm ratio of TFEB‐mNG in non‐treated (nt) or LLOMe treated cells shown in (E). ≥ 80 cells were analyzed per experiment for each condition.
Representative immunoblot of EGFR in siSTK38‐transfected HeLa cells. Cells were treated with EGF (50 ng/ml for the indicated number of minutes).
Quantification of EGFR shown in (G).

Schematic diagram of EGFP‐TRPML1 cleavage assay.
Representative immunoblots of EGFP‐TRPML1 in MCF10A cells transfected with siRNAs of ESCRT components. Cells were treated with nigericin (50 μM for 8 h).
Quantification of cleaved EGFP shown in (B).
Representative immunoblots of EGFP‐TRPML1 in siSTK38‐transfected WT or ATG13 KO MCF10A cells. Cells were treated with nigericin (50 μM for 8 h). The asterisk in STK38 blot represents non‐specific band.
Quantification of cleaved EGFP shown in (D).
Representative immunoblots of EGFP‐TRPML1 in siSTK38‐transfected MCF10A cells. Cells were treated with LLOMe (0.5 mM for 1 h, then incubated for 7 h after washout).
Quantification of cleaved EGFP shown in (F).
Representative images of multivesicular structures (arrows) obtained by transmission electron microscopy (TEM). siSTK38‐transfected MCF10A cells were treated with NH4Cl (5 mM for 24 h). Scale bars: 1 μm.
Quantification of ILVs per multivesicular structure shown in (H). ≥ 20 multivesicular structures from 10 independent cells were analyzed per condition.
Representative images of CLEM analysis of LAMP1‐GFP positive endolysosomes. LAMP1‐GFP expressing MCF10A cells were treated with NH4Cl (5 mM for 24 h). Scale bars: 10 μm (low magnification images) or 1 μm (magnified images).

Representative immunoblots of EGFP‐TRPML1 in WT, ATG5 KO, and ATG13 KO MCF10A cells. Cells were treated with monensin (10 μM), nigericin (5 μM), or LLOMe (0.5 mM) for 8 h.
Quantification of cleaved EGFP shown in (A).
Representative images of macroautophagy‐related factors (LC3, Gal3, ubiquitin‐K48 chain (Ub‐K48), and p62, shown in green), LAMP1 (magenta), and DAPI (blue) in MCF10A cells. Cells were treated with LLOMe (0.5 mM for 1 h), nigericin (5 μM for 2 h), monensin (10 μM for 2 h), or NH4Cl (5 mM for 2 h). Scale bars: 10 μm.
Representative images of stably expressed EGFP‐TRPML1 (green), immunostained LAMP1 (magenta), and DAPI (blue) in MCF10A cells. Cells were treated with monensin (50 μM) for 1 h, followed by the co‐treatment with Apilimod (200 nM) for 2 h. Scale bars: 10 μm.
Representative images of stably expressed EGFP‐TRPML1 (green), immunostained CD63 (magenta), and DAPI (blue) in MCF10A cells. Cells were treated with monensin (50 μM) for 1 h, followed by the co‐treatment with Apilimod (200 nM) for 2 h. Scale bars: 10 μm.
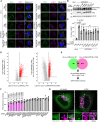
Representative images of immunostained VPS4 (green), LAMP1 (magenta), and DAPI (blue) in STK38 reconstituted HeLa cells. Cells were transfected with siSTK38 for 48 h, then transfected with Flag‐tagged STK38. After 24 h of plasmid transfection, cells were treated with LLOMe (1 mM for 30 min). Scale bars: 10 μm.
Representative immunoblots of STK38 in Flag‐STK38 reconstituted HeLa cells in (A). The asterisk in STK38 blot represents STK38L.
Quantification of co‐localization between VPS4 and LAMP1 shown in (A). ≥ 50 cells were analyzed per experiment for each condition.
Volcano plots of tandem mass tag phosphoproteomics data. Phosphopeptides from siLuciferace (Luc)‐transfected HeLa cells treated with LLOMe (siLuc‐LLOMe) were compared with siLuc‐transfected non‐treated cells (siLuc‐NT) (left) or with siSTK38‐transfected cells treated with LLOMe (siSTK38‐LLOMe) (right). The horizontal line indicates the significance cutoff (adjusted P‐value < 0.05). The vertical line indicates the fold‐change cutoff (FC ≥ 1.6). Red dots indicate phosphopeptides above the significance and fold‐change thresholds. The data was obtained from three independent experiments.
Schematic diagram of candidate selection based on phosphoproteomics results. The Venn diagram represents the number of phosphopeptides above the thresholds from the two comparisons shown in (D). From 180 phosphopeptides, we selected 25 candidates that shared a common AGC kinase phosphorylation motif (R/K‐x‐x‐pS/pT).
Quantification of VPS4 dots in HeLa cells transfected with siRNAs of 25 candidates. Cells were treated with LLOMe (1 mM for 30 min). ≥ 70 cells were analyzed per experiment for each condition.
Representative images of transiently expressed mNG‐DOK1 (green) and immunostained LAMP1 (magenta) in HeLa cells treated with LLOMe (1 mM for 1 h, then incubated for 3 h after washout). Scale bars: 10 μm.
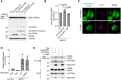
Representative immunoblots of EGFP‐TRPML1 in MCF10A cells expressing HA tag only, WT HA‐STK38, or kinase‐dead HA‐STK38 (K118R). Cells were treated with nigericin (50 μM for 8 h).
Quantification of cleaved EGFP shown in (A).
Representative images of transiently expressed kinase‐dead K118R mutant of mNG‐STK38 (green) and immunostained LAMP1 (magenta) in HeLa cells treated with LLOMe (1 mM for 1 h, then incubated for 3 h after washout). Scale bars: 10 μm.
Relative expression levels of DOK1 mRNA in DOK1‐reconstituted HeLa cells shown in Fig 5C and D.
Representative immunoblots from a co‐immunoprecipitation experiment. HeLa cells expressing Flag‐DOK1 were treated with LLOMe (1 mM for 30 min). Cell lysates were immunoprecipitated using anti‐Flag‐beads agarose.
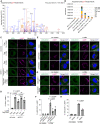
Phosphorylation of DOK1 S269 was demonstrated by the MS/MS spectrum of the m/z 561.25214 ion from siLuc‐transfected and LLOMe‐treated cells.
PRM‐based quantification of the phosphopeptide shown in (A). HeLa cells stably expressing mNG‐DOK1 were transfected with siLuc or siSTK38. Then, cells were treated with LLOMe (1 mM) for (i) 30 min or (ii) 1 h plus incubation for 5 h after washout (1 h + wo 5 h).
Representative images of immunostained VPS4 (green), LAMP1 (magenta), and DAPI (blue) in DOK1 reconstituted HeLa cells. Cells were transfected with siDOK1 for 48 h, then transfected with Flag‐tagged DOK1. After 24 h of plasmid transfection, cells were treated with LLOMe (1 mM for 30 min). Scale bars: 10 μm.
Quantification of co‐localization between VPS4 and LAMP1 shown in (C). ≥ 50 cells were analyzed per experiment for each condition.
Representative images of stably expressed mNG‐DOK1 (green), LAMP1 (magenta), and DAPI (blue) in siSTK38‐transfected HeLa cells. Cells were treated with LLOMe (1 mM for 1 h). Scale bars: 10 μm.
Quantification of co‐localization between mNG‐DOK1 and LAMP1 shown in (E). ≥ 50 cells were analyzed per experiment for each condition.
Representative images of transiently expressed WT or S269A mNG‐DOK1 (green), LAMP1 (magenta), and DAPI (blue) in HeLa cells. Cells were treated with LLOMe (1 mM for 1 h). Scale bars: 10 μm.
Quantification of co‐localization between mNG‐DOK1 and LAMP1 shown in (G). ≥ 50 cells were analyzed per experiment for each condition.
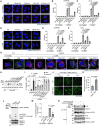
Representative images of immunostained CHMP4B (green), ALIX (magenta), and DAPI (blue) in WT, FIP200, ATG3, ATG7, and ATG16L1 KO HeLa cells. Cells were treated with LLOMe (1 mM for 30 min). Scale bars: 10 μm.
Quantification of CHMP4B dots shown in (A). ≥ 100 cells were analyzed per experiment for each condition.
Quantification of ALIX dots shown in (A). ≥ 100 cells were analyzed per experiment for each condition.
Representative images of immunostained CHMP4B (green), ALIX (magenta), and DAPI (blue) in WT, ATG8 hexa KO, LC3 TKO, and GABARAP (GBRP) TKO HeLa cells. Cells were treated with LLOMe (1 mM for 30 min). Scale bars: 10 μm.
Quantification of CHMP4B dots shown in (D). ≥ 100 cells were analyzed per experiment for each condition.
Quantification of ALIX dots shown in (D). ≥ 100 cells were analyzed per experiment for each condition.
Representative images of immunostained CHMP4B (green), LAMP1 (magenta), and DAPI (blue) in WT and ATG8 hexa KO HeLa cells expressing 3×Flag‐ATG8 paralogs. Cells were treated with LLOMe (1 mM for 30 min). Scale bars: 10 μm.
Representative immunoblots of Flag‐ATG8s expressed in HeLa cells.
Quantification of co‐localization between CHMP4B and LAMP1 shown in (G). ≥ 100 cells were analyzed per experiment for each condition.
Representative images of stably expressed GFP‐Gal3 (green) in WT or GABARAP TKO HeLa cells. Cells were treated with LLOMe (1 mM for 1 h, then incubated for the indicated number of hours after washout). Scale bars: 10 μm.
Percentage of residual GFP‐Gal3 dots (10 h/0 h [%]) in (J). ≥ 100 cells were analyzed per experiment for each condition.
Representative immunoblots of EGFP‐TRPML1 and GABARAPs in MCF10A cells transfected with siGABARAP (siRNAs of GABARAP, GABARAPL1, and GABARAPL2). Cells were treated with nigericin (50 μM for 8 h). The asterisk in GABARAPL1 blot represents a non‐specific band.
Quantification of cleaved EGFP shown in (L).
Representative immunoblots from a co‐immunoprecipitation experiment. mCherry‐LC3B‐ or GABARAPL2‐expressing hexa KO HeLa cells were treated with LLOMe (1 mM for 30 min). Cell lysates were immunoprecipitated using RFP‐trap.
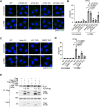
Representative images of immunostained VPS4 (green) and DAPI (blue) in WT, FIP200, ATG3, ATG7, and ATG16L1 KO HeLa cells. Cells were treated with LLOMe (1 mM for 30 min). Scale bars: 10 μm.
Quantification of VPS4 dots shown in (A). ≥ 100 cells were analyzed per experiment for each condition.
Representative images of immunostained VPS4 (green) and DAPI (blue) in WT, ATG8 hexa KO, LC3 TKO, and GABARAP TKO HeLa cells. Cells were treated with LLOMe (1 mM for 30 min). Scale bars: 10 μm.
Quantification of VPS4 dots shown in (C). ≥ 100 cells were analyzed per experiment for each condition.
Representative immunoblots from a co‐immunoprecipitation experiment. Flag‐LC3B‐ or GABARAPL2‐expressing hexa KO HeLa cells were treated with LLOMe (1 mM for 30 min). Cell lysates were immunoprecipitated using anti‐Flag‐beads agarose. Fold changes in co‐immunoprecipitated ALIX were quantified (Fold change vs. non‐treated Flag‐LC3B, normalized to Flag). The asterisk in ALIX blot represents a non‐specific band.
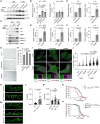
Representative immunoblots of p21 and p16 in siSTK38‐transfected senescent (+ DXR) or non‐induced (− DXR) RPE1 cells. Cellular senescence was induced by doxorubicin for 3 days.
Quantification of p21 and p16 in senescent induced cells shown in (A).
Representative immunoblots of p21 and p16 in siGABARAP‐transfected senescent (+ DXR) or non‐induced (− DXR) RPE1 cells. Cellular senescence was induced by doxorubicin for 3 days.
Quantification of p21 and p16 in senescent induced cells shown in (C).
Relative expression levels of IL6 and IL1A mRNA in siSTK38‐transfected senescent RPE1 cells. Cellular senescence was induced by doxorubicin for 3 days.
Relative expression levels of IL6 and IL1A mRNA in siGABARAP‐transfected senescent RPE1 cells. Cellular senescence was induced by doxorubicin for 3 days.
Representative images of SA‐β‐Gal–positive cells in siSTK38‐ or siGABARAP‐transfected senescent RPE1 cells. Cellular senescence was induced by doxorubicin for 3 days. Scale bars: 100 μm.
Percentage of SA‐β‐Gal–positive cells in (G). ≥ 500 cells were analyzed per experiment for each condition.
Representative images of stably expressed GFP‐Gal3 (green) and immunostainied LAMP1 (magenta) in siSTK38‐ or siGABARAP‐transfected senescent RPE1 cells. Cellular senescence was induced by doxorubicin for 10 days. Scale bars: 10 μm.
Quantification of co‐localization between GFP‐Gal3 and LAMP1 shown in (I). ≥ 100 cells were analyzed for each condition from 5 independent experiments.
Representative images of sax‐1 KO C. elegans expressing GFP‐Gal3 at day 7 of the adult stage. Scale bars: 20 μm.
Quantification of GFP‐Gal3 dots shown in (K). ≥ 29 worms were analyzed for each condition from 3 independent experiments.
Representative images of lgg‐1 or lgg‐2 knocked down (KD) C. elegans expressing GFP‐Gal3 at day 7 of the adult stage. Scale bars: 20 μm.
Quantification of GFP‐Gal3 dots shown in (M). ≥ 29 worms were analyzed for each condition from 3 independent experiments.
Lifespan curves of N2 (WT) and sax‐1 KO worms.
Lifespan curves of N2 (WT), lgg‐1 KD, and lgg‐2 KD worms.
References
-
- Ballabio A, Bonifacino JS (2020) Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol 21: 101–118 - PubMed
-
- Brummer T, Schmitz‐Peiffer C, Daly RJ (2010) Docking proteins. FEBS J 277: 4356–4369 - PubMed
-
- Carpino N, Wisniewski D, Strife A, Marshak D, Kobayashi R, Stillman B, Clarkson B (1997) p62(dok): a constitutively tyrosine‐phosphorylated, GAP‐associated protein in chronic myelogenous leukemia progenitor cells. Cell 88: 197–204 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Astellas Foundation for Research on Metabolic Disorders
- JP22gm1410014/Japan Agency for Medical Research and Development (AMED)
- 21J21647/MEXT | Japan Society for the Promotion of Science (JSPS)
- 22H04982/MEXT | Japan Society for the Promotion of Science (JSPS)
- 21H02428/MEXT | Japan Society for the Promotion of Science (JSPS)
- 23K18140/MEXT | Japan Society for the Promotion of Science (JSPS)
- JPMJCR17H6/MEXT | JST | Core Research for Evolutional Science and Technology (CREST)
- 21H05145/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
- Mitsubishi Foundation (The Mitsubishi Foundation)
- Mochida Memorial Foundation for Medical and Pharmaceutical Research ( )
- NOVARTIS Foundation (Japan) for the Promotion of Science (NOVARTIS Foundation (Japan))
- Takeda Science Foundation (TSF)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

