Topoisomerase 1 Inhibition in MYC-Driven Cancer Promotes Aberrant R-Loop Accumulation to Induce Synthetic Lethality
- PMID: 37987734
- PMCID: PMC10722143
- DOI: 10.1158/0008-5472.CAN-22-2948
Topoisomerase 1 Inhibition in MYC-Driven Cancer Promotes Aberrant R-Loop Accumulation to Induce Synthetic Lethality
Abstract
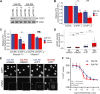
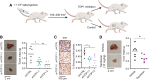
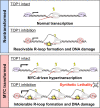
MYC is a central regulator of gene transcription and is frequently dysregulated in human cancers. As targeting MYC directly is challenging, an alternative strategy is to identify specific proteins or processes required for MYC to function as a potent cancer driver that can be targeted to result in synthetic lethality. To identify potential targets in MYC-driven cancers, we performed a genome-wide CRISPR knockout screen using an isogenic pair of breast cancer cell lines in which MYC dysregulation is the switch from benign to transformed tumor growth. Proteins that regulate R-loops were identified as a potential class of synthetic lethal targets. Dysregulated MYC elevated global transcription and coincident R-loop accumulation. Topoisomerase 1 (TOP1), a regulator of R-loops by DNA topology, was validated to be a vulnerability in cells with high MYC activity. Genetic knockdown of TOP1 in MYC-transformed cells resulted in reduced colony formation compared with control cells, demonstrating synthetic lethality. Overexpression of RNaseH1, a riboendonuclease that specifically degrades R-loops, rescued the reduction in clonogenicity induced by TOP1 deficiency, demonstrating that this vulnerability is driven by aberrant R-loop accumulation. Genetic and pharmacologic TOP1 inhibition selectively reduced the fitness of MYC-transformed tumors in vivo. Finally, drug response to TOP1 inhibitors (i.e., topotecan) significantly correlated with MYC levels and activity across panels of breast cancer cell lines and patient-derived organoids. Together, these results highlight TOP1 as a promising target for MYC-driven cancers.
Significance: CRISPR screening reveals topoisomerase 1 as an immediately actionable vulnerability in cancers harboring MYC as a driver oncoprotein that can be targeted with clinically approved inhibitors.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures




References
-
- Lourenco C, Resetca D, Redel C, Lin P, MacDonald AS, Ciaccio R, et al. . MYC protein interactors in gene transcription and cancer. Nat Rev Cancer 2021;21:579–91. - PubMed
-
- Thng DKH, Toh TB, Chow EK. Capitalizing on synthetic lethality of MYC to treat cancer in the digital age. Trends Pharmacol Sci 2021;42:166–82. - PubMed

