Editing the 19 kDa alpha-zein gene family generates non-opaque2-based quality protein maize
- PMID: 37988568
- PMCID: PMC10955486
- DOI: 10.1111/pbi.14237
Editing the 19 kDa alpha-zein gene family generates non-opaque2-based quality protein maize
Abstract
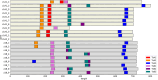
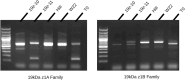
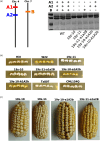
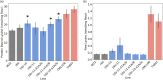
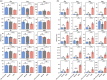
Maize grain is deficient in lysine. While the opaque2 mutation increases grain lysine, o2 is a transcription factor that regulates a wide network of genes beyond zeins, which leads to pleiotropic and often negative effects. Additionally, the drastic reduction in 19 kDa and 22 kDa alpha-zeins causes a floury kernel, unsuitable for agricultural use. Quality protein maize (QPM) overcame the undesirable kernel texture through the introgression of modifying alleles. However, QPM still lacks a functional o2 transcription factor, which has a penalty on non-lysine amino acids due to the o2 mutation. CRISPR/cas9 gives researchers the ability to directly target genes of interest. In this paper, gene editing was used to specifically target the 19 kDa alpha zein gene family. This allows for proteome rebalancing to occur without an o2 mutation and without a total alpha-zein knockout. The results showed that editing some, but not all, of the 19 kDa zeins resulted in up to 30% more lysine. An edited line displayed an increase of 30% over the wild type. While not quite the 55% lysine increase displayed by QPM, the line had little collateral impact on other amino acid levels compared to QPM. Additionally, the edited line containing a partially reduced 19 kDa showed an advantage in kernel texture that had a complete 19 kDa knockout. These results serve as proof of concept that editing the 19 kDa alpha-zein family alone can enhance lysine while retaining vitreous endosperm and a functional O2 transcription factor.
Keywords: CRISPR/cas9; QPM; lysine; opaque2; seed storage proteins; zeins.
© 2023 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Armstrong, C. (1991) Development and availability of germplasm with high type ii culture response. Maize Genet. Coop. News Lett. 65, 92–93.
-
- Bressani, R. , Benavides, V. , Acevedo, E. , Ortiz, M. et al. (1990) Changes in selected nutrient contents and in protein quality of common and quality protein maize during rural tortilla preparation. Cereal. Chem. 67, 515–518.
-
- Burgoon, K. , Hansen, J. , Knabe, D. and Bockholt, A. (1992) Nutritional value of quality protein maize for starter and finisher swine. J. Anim. Sci. 70, 811–817. - PubMed
-
- Dai, D. , Ma, Z. and Song, R. (2021) Maize endosperm development. J. Integr. Plant Biol. 63, 613–627. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

