Insight into the on/off switch that regulates expression of the MSMEG-3762/63 efflux pump in Mycobacterium smegmatis
- PMID: 37989843
- PMCID: PMC10663510
- DOI: 10.1038/s41598-023-47695-4
Insight into the on/off switch that regulates expression of the MSMEG-3762/63 efflux pump in Mycobacterium smegmatis
Abstract
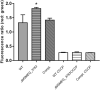
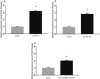
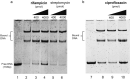
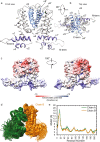
Drug resistance is one of the most difficult challenges facing tuberculosis (TB) control. Drug efflux is among the mechanisms leading to drug resistance. In our previous studies, we partially characterized the ABC-type MSMEG-3762/63 efflux pump in Mycobacterium smegmatis, which shares high percentage of identity with the Mycobacterium tuberculosis Rv1687/86c pump. MSMEG-3762/63 was shown to have extrusion activity for rifampicin and ciprofloxacin, used in first and second-line anti-TB treatments. Moreover, we described the functional role of the TetR-like MSMEG-3765 protein as a repressor of the MSMEG_3762/63/65 operon and orthologous Rv1687/86/85c in M. tuberculosis. Here we show that the operon is upregulated in the macrophage environment, supporting a previous observation of induction triggered by acid-nitrosative stress. Expression of the efflux pump was also induced by sub-inhibitory concentrations of rifampicin or ciprofloxacin. Both these drugs also prevented the binding of the MSMEG-3765 TetR repressor protein to its operator in the MSMEG_3762/63/65 operon. The hypothesis that these two drugs might be responsible for the induction of the efflux pump operon was assessed by bioinformatics analyses. Docking studies using a structural model of the regulator MSMEG-3765 showed that both antibiotics abolished the ability of this transcriptional repressor to recognize the efflux pump operon by interacting with the homodimer at different binding sites within the same binding pocket. Reduced binding of the repressor leads to induction of the efflux pump in M. smegmatis, and reduced efficacy of these two anti-mycobacterial drugs.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Characterization of the Mycobacterial MSMEG-3762/63 Efflux Pump in Mycobacterium smegmatis Drug Efflux.Front Microbiol. 2020 Dec 3;11:575828. doi: 10.3389/fmicb.2020.575828. eCollection 2020. Front Microbiol. 2020. PMID: 33343518 Free PMC article.
-
A Novel TetR-Like Transcriptional Regulator Is Induced in Acid-Nitrosative Stress and Controls Expression of an Efflux Pump in Mycobacteria.Front Microbiol. 2017 Oct 23;8:2039. doi: 10.3389/fmicb.2017.02039. eCollection 2017. Front Microbiol. 2017. PMID: 29109706 Free PMC article.
-
Ajoene: a natural compound with enhanced antimycobacterial and antibiofilm properties mediated by efflux pump modulation and ROS generation against M. Smegmatis.Arch Microbiol. 2024 Nov 2;206(12):453. doi: 10.1007/s00203-024-04189-9. Arch Microbiol. 2024. PMID: 39487375
-
Mycobacterium Lrp/AsnC family transcriptional factor modulates the arginase pathway as both a sensor and a transcriptional repressor.J Genet Genomics. 2021 Nov 20;48(11):1020-1031. doi: 10.1016/j.jgg.2021.06.018. Epub 2021 Jul 29. J Genet Genomics. 2021. PMID: 34696992 Review.
-
Energy metabolism and drug efflux in Mycobacterium tuberculosis.Antimicrob Agents Chemother. 2014 May;58(5):2491-503. doi: 10.1128/AAC.02293-13. Epub 2014 Mar 10. Antimicrob Agents Chemother. 2014. PMID: 24614376 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

