Systemic perturbations in amino acids/amino acid derivatives and tryptophan pathway metabolites associated with murine influenza A virus infection
- PMID: 37990229
- PMCID: PMC10664681
- DOI: 10.1186/s12985-023-02239-0
Systemic perturbations in amino acids/amino acid derivatives and tryptophan pathway metabolites associated with murine influenza A virus infection
Abstract
Background: Influenza A virus (IAV) is the only influenza virus causing flu pandemics (i.e., global epidemics of flu disease). Influenza (the flu) is a highly contagious disease that can be deadly, especially in high-risk groups. Worldwide, these annual epidemics are estimated to result in about 3 to 5 million cases of severe illness and in about 290,000 to 650,000 respiratory deaths. We intend to reveal the effect of IAV infection on the host's metabolism, immune response, and neurotoxicity by using a mouse IAV infection model.
Methods: 51 metabolites of murine blood plasma (33 amino acids/amino acid derivatives (AADs) and 18 metabolites of the tryptophan pathway) were analyzed by using Ultra-High-Performance Liquid Chromatography-Mass Spectrometry with Electrospray Ionization at the acute (7 days post-infection (dpi)), resolution (14 dpi), and recovery (21 dpi) stages of the virus infection in comparison with controls.
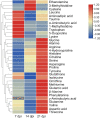
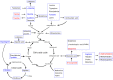
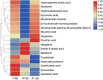
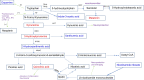
Results: Among the 33 biogenic amino acids/AADs, the levels of five amino acids/AADs (1-methylhistidine, 5-oxoproline, α-aminobutyric acid, glutamine, and taurine) increased by 7 dpi, whereas the levels of ten amino acids/AADs (4-hydroxyproline, alanine, arginine, asparagine, cysteine, citrulline, glycine, methionine, proline, and tyrosine) decreased. By 14 dpi, the levels of one AAD (3-methylhistidine) increased, whereas the levels of five amino acids/AADs (α-aminobutyric acid, aminoadipic acid, methionine, threonine, valine) decreased. Among the 18 metabolites from the tryptophan pathway, the levels of kynurenine, quinolinic acid, hydroxykynurenine increased by 7 dpi, whereas the levels of indole-3-acetic acid and nicotinamide riboside decreased.
Conclusions: Our data may facilitate understanding the molecular mechanisms of host responses to IAV infection and provide a basis for discovering potential new mechanistic, diagnostic, and prognostic biomarkers and therapeutic targets for IAV infection.
Keywords: Amino acids; Immunometabolism; Infection; Influenza; Influenza A virus; Metabolites; Neurotoxicity; Tryptophan pathway.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
AMINO ACID COMPOSITION OF HIGHLY PURIFIED VIRAL PARTICLES OF INFLUENZA A AND B.J Exp Med. 1947 Jul 31;86(2):125-9. doi: 10.1084/jem.86.2.125. J Exp Med. 1947. PMID: 19871660 Free PMC article.
-
Targeted Amino Acids Profiling of Human Seminal Plasma from Teratozoospermia Patients Using LC-MS/MS.Reprod Sci. 2023 Nov;30(11):3285-3295. doi: 10.1007/s43032-023-01272-2. Epub 2023 Jun 1. Reprod Sci. 2023. PMID: 37264261
-
A Pilot Study for Investigation of Plasma Amino Acid Profile in Neurofibromatosis Type 1 Patients.Comb Chem High Throughput Screen. 2022;25(1):114-122. doi: 10.2174/1386207323666201204143206. Comb Chem High Throughput Screen. 2022. PMID: 33280590
-
Impact of Reactive Species on Amino Acids-Biological Relevance in Proteins and Induced Pathologies.Int J Mol Sci. 2022 Nov 14;23(22):14049. doi: 10.3390/ijms232214049. Int J Mol Sci. 2022. PMID: 36430532 Free PMC article. Review.
-
Innate and adaptive immune responses against Influenza A Virus: Immune evasion and vaccination strategies.Immunobiology. 2022 Nov;227(6):152279. doi: 10.1016/j.imbio.2022.152279. Epub 2022 Sep 14. Immunobiology. 2022. PMID: 36272344 Review.
Cited by
-
Multiomics analysis unveils key biomarkers during dynamic progress of IAV infection in mice.Front Immunol. 2025 May 22;16:1566690. doi: 10.3389/fimmu.2025.1566690. eCollection 2025. Front Immunol. 2025. PMID: 40475788 Free PMC article.
-
Impaired arginine/ornithine metabolism drives severe HFMD by promoting cytokine storm.Front Immunol. 2024 Jun 24;15:1407035. doi: 10.3389/fimmu.2024.1407035. eCollection 2024. Front Immunol. 2024. PMID: 38979420 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

