Comparison of new computational methods for spatial modelling of malaria
- PMID: 37990242
- PMCID: PMC10664662
- DOI: 10.1186/s12936-023-04760-7
Comparison of new computational methods for spatial modelling of malaria
Abstract
Background: Geostatistical analysis of health data is increasingly used to model spatial variation in malaria prevalence, burden, and other metrics. Traditional inference methods for geostatistical modelling are notoriously computationally intensive, motivating the development of newer, approximate methods for geostatistical analysis or, more broadly, computational modelling of spatial processes. The appeal of faster methods is particularly great as the size of the region and number of spatial locations being modelled increases.
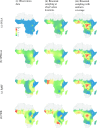
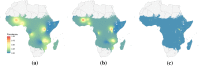
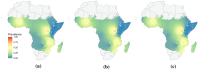
Methods: This work presents an applied comparison of four proposed 'fast' computational methods for spatial modelling and the software provided to implement them-Integrated Nested Laplace Approximation (INLA), tree boosting with Gaussian processes and mixed effect models (GPBoost), Fixed Rank Kriging (FRK) and Spatial Random Forests (SpRF). The four methods are illustrated by estimating malaria prevalence on two different spatial scales-country and continent. The performance of the four methods is compared on these data in terms of accuracy, computation time, and ease of implementation.
Results: Two of these methods-SpRF and GPBoost-do not scale well as the data size increases, and so are likely to be infeasible for larger-scale analysis problems. The two remaining methods-INLA and FRK-do scale well computationally, however the resulting model fits are very sensitive to the user's modelling assumptions and parameter choices. The binomial observation distribution commonly used for disease prevalence mapping with INLA fails to account for small-scale overdispersion present in the malaria prevalence data, which can lead to poor predictions. Selection of an appropriate alternative such as the Beta-binomial distribution is required to produce a reliable model fit. The small-scale random effect term in FRK overcomes this pitfall, but FRK model estimates are very reliant on providing a sufficient number and appropriate configuration of basis functions. Unfortunately the computation time for FRK increases rapidly with increasing basis resolution.
Conclusions: INLA and FRK both enable scalable geostatistical modelling of malaria prevalence data. However care must be taken when using both methods to assess the fit of the model to data and plausibility of predictions, in order to select appropriate model assumptions and parameters.
Keywords: Geostatistics; Predictive modelling; Risk mapping; Spatial modelling.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



















References
-
- Diggle P, Ribeiro Jr PJ. Model-based geostatistics. Springer; 2007.
-
- Martínez-Minaya J, Cameletti M, Conesa D, Pennino MG. Species distribution modelling: a statistical review with focus in spatio-temporal issues. Stoch Environ Res Risk Assess. 2018;32:3227–3244. doi: 10.1007/s00477-018-1548-7. - DOI
-
- Holdaway MR. Spatial modelling and interpolation of monthly temperature using kriging. Clim Res. 1996;6(3):215–225. doi: 10.3354/cr006215. - DOI
-
- Samalot A, Astitha M, Yang J, Galanis G. Combined Kalman filter and universal kriging to improve storm wind speed predictions for the northeastern United States. Weather Forecast. 2019;34(3):587–601. doi: 10.1175/WAF-D-18-0068.1. - DOI
-
- Mulla D. Mapping and managing spatial patterns in soil fertility and crop yield. In: Proceedings of soil specific crop management: a workshop on research and development issues. Wiley Online Library; 1993. pp. 15–26.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

