HIV-2 inhibits HIV-1 gene expression via two independent mechanisms during cellular co-infection
- PMID: 37991365
- PMCID: PMC10734542
- DOI: 10.1128/jvi.01870-22
HIV-2 inhibits HIV-1 gene expression via two independent mechanisms during cellular co-infection
Abstract
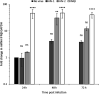
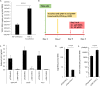
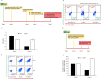
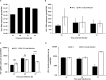
Twenty-five years after the first report that HIV-2 infection can reduce HIV-1-associated pathogenesis in dual-infected patients, the mechanisms are still not well understood. We explored these mechanisms in cell culture and showed first that these viruses can co-infect individual cells. Under specific conditions, HIV-2 inhibits HIV-1 through two distinct mechanisms, a broad-spectrum interferon response and an HIV-1-specific inhibition conferred by the HIV-2 TAR. The former could play a prominent role in dually infected individuals, whereas the latter targets HIV-1 promoter activity through competition for HIV-1 Tat binding when the same target cell is dually infected. That mechanism suppresses HIV-1 transcription by stalling RNA polymerase II complexes at the promoter through a minimal inhibitory region within the HIV-2 TAR. This work delineates the sequence of appearance and the modus operandi of each mechanism.
Keywords: HIV-1; HIV-2; HIV-2 TAR; RNA polymerase II; dual infection; interferon response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Barré-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vézinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868–871. doi:10.1126/science.6189183 - DOI - PubMed
-
- WHO . 2021. HIV/AIDS. World Health Organization.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

