De novo assembly and comparative genome analysis for polyhydroxyalkanoates-producing Bacillus sp. BNPI-92 strain
- PMID: 37991636
- PMCID: PMC10665291
- DOI: 10.1186/s43141-023-00578-7
De novo assembly and comparative genome analysis for polyhydroxyalkanoates-producing Bacillus sp. BNPI-92 strain
Abstract
Background: Certain Bacillus species play a vital role in polyhydroxyalkanoate (PHA) production. However, most of these isolates did not properly identify to species level when scientifically had been reported.
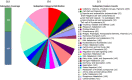
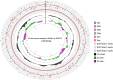
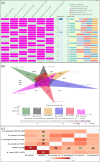
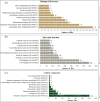
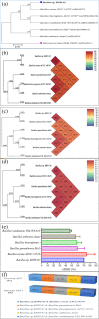
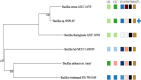
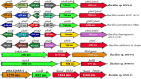
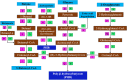
Results: From NGS analysis, 5719 genes were predicted in the de novo genome assembly. Based on genome annotation using RAST server, 5,527,513 bp sequences were predicted with 5679 bp number of protein-coding sequence. Its genome sequence contains 35.1% and 156 GC content and contigs, respectively. In RAST server analysis, subsystem (43%) and non-subsystem coverage (57%) were generated. Ortho Venn comparative genome analysis indicated that Bacillus sp. BNPI-92 shared 2930 gene cluster (core gene) with B. cereus ATCC 14579 T (AE016877), B. paranthracis Mn5T (MACE01000012), B. thuringiensis ATCC 10792 T (ACNF01000156), and B. antrics Amen T (AE016879) strains. For our strain, the maximum gene cluster (190) was shared with B. cereus ATCC 14579 T (AE016877). For Ortho Venn pair wise analysis, the maximum overlapping gene clusters thresholds have been detected between Bacillus s p.BNPI-92 and Ba. cereus ATCC 14579 T (5414). Average nucleotide identity (ANI) such as OriginalANI and OrthoANI, in silicon digital DND-DNA hybridization (isDDH), Type (Strain) Genome Server (TYGS), and Genome-Genome Distance Calculator (GGDC) were more essentially related Bacillus sp. BNPI-92 with B. cereus ATCC 14579 T strain. Therefore, based on the combination of RAST annotation, OrthoVenn server, ANI and isDDH result Bacillus sp.BNPI-92 strain was strongly confirmed to be a B. cereus type strain. It was designated as B. cereus BNPI-92 strain. In B. cereus BNPI-92 strain whole genome sequence, PHA biosynthesis encoding genes such as phaP, phaQ, phaR (PHA synthesis repressor phaR gene sequence), phaB/phbB, and phaC were predicted on the same operon. These gene clusters were designated as phaPQRBC. However, phaA was located on other operons.
Conclusions: This newly obtained isolate was found to be new a strain based on comparative genomic analysis and it was also observed as a potential candidate for PHA biosynthesis.
Keywords: ANI; Bacillus species; Gene; Genome DNA; Metabolic pathway.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests. Ethiopian Embassy, Adama Science and Technology University, and KIIT University who funded this project had no role in the design of the proposal development, the study design, sample collection, data analyses, and data interpretation, in the writing of this manuscript, or in the decision to publish the current results.
Figures

Similar articles
-
Optimization of the culture conditions for production of Polyhydroxyalkanoate and its characterization from a new Bacillus cereus sp. BNPI-92 strain, isolated from plastic waste dumping yard.Int J Biol Macromol. 2020 Aug 1;156:1064-1080. doi: 10.1016/j.ijbiomac.2019.11.138. Epub 2019 Nov 18. Int J Biol Macromol. 2020. PMID: 31751740
-
Novel Effective Bacillus cereus Group Species "Bacillus clarus" Is Represented by Antibiotic-Producing Strain ATCC 21929 Isolated from Soil.mSphere. 2020 Nov 4;5(6):e00882-20. doi: 10.1128/mSphere.00882-20. mSphere. 2020. PMID: 33148822 Free PMC article.
-
Complete genome sequence of novel isolate SYJ15 of Bacillus cereus group, a highly lethal pathogen isolated from Chinese soft shell turtle (Pelodiscus Sinensis).Arch Microbiol. 2020 Jan;202(1):85-92. doi: 10.1007/s00203-019-01723-y. Epub 2019 Sep 4. Arch Microbiol. 2020. PMID: 31485709
-
Description of Bacillus toyonensis sp. nov., a novel species of the Bacillus cereus group, and pairwise genome comparisons of the species of the group by means of ANI calculations.Syst Appl Microbiol. 2013 Sep;36(6):383-91. doi: 10.1016/j.syapm.2013.04.008. Epub 2013 Jun 19. Syst Appl Microbiol. 2013. PMID: 23791203
-
Class IV polyhydroxyalkanoate (PHA) synthases and PHA-producing Bacillus.Appl Microbiol Biotechnol. 2015 Aug;99(15):6231-40. doi: 10.1007/s00253-015-6777-9. Epub 2015 Jul 2. Appl Microbiol Biotechnol. 2015. PMID: 26135986 Review.
Cited by
-
Two colistin resistance-producing Aeromonas strains, isolated from coastal waters in Zhejiang, China: characteristics, multi-drug resistance and pathogenicity.Front Microbiol. 2024 Jul 31;15:1401802. doi: 10.3389/fmicb.2024.1401802. eCollection 2024. Front Microbiol. 2024. PMID: 39144207 Free PMC article.
-
Environmental impact on the genome shaping of putative new Streptomyces species.BMC Microbiol. 2025 Feb 12;25(1):72. doi: 10.1186/s12866-025-03779-x. BMC Microbiol. 2025. PMID: 39939924 Free PMC article.
-
Novel Anti-MRSA Peptide from Mangrove-Derived Virgibacillus chiguensis FN33 Supported by Genomics and Molecular Dynamics.Mar Drugs. 2025 May 14;23(5):209. doi: 10.3390/md23050209. Mar Drugs. 2025. PMID: 40422799 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
