Rapidly determining the 3D structure of proteins by surface-enhanced Raman spectroscopy
- PMID: 37992170
- PMCID: PMC10665000
- DOI: 10.1126/sciadv.adh8362
Rapidly determining the 3D structure of proteins by surface-enhanced Raman spectroscopy
Abstract
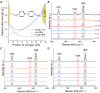
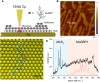
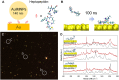
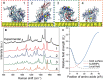
Despite great advances in protein structure analysis, label-free and ultrasensitive methods to obtain the natural and dynamic three-dimensional (3D) structures are still urgently needed. Surface-enhanced Raman spectroscopy (SERS) can be a good candidate, whereas the complexity originated from the interactions between the protein and the gradient surface electric field makes it extremely challenging to determine the protein structure. Here, we propose a deciphering strategy for accurate determination of 3D protein structure from experimental SERS spectra in seconds by simply summing SERS spectra of isolated amino acids in electric fields of different strength with their orientations in protein. The 3D protein structure can be reconstructed by comparing the experimental spectra obtained in a well-defined gap-mode SERS configuration with the simulated spectra. The gradient electric field endows SERS with a unique advantage to section biomolecules with atomic precision, which makes SERS a competent tool for monitoring biomolecular events under physiological conditions.
Figures

References
-
- Cao L., Coventry B., Goreshnik I., Huang B., Sheffler W., Park J. S., Jude K. M., Marković I., Kadam R. U., Verschueren K. H. G., Verstraete K., Walsh S. T. R., Bennett N., Phal A., Yang A., Kozodoy L., DeWitt M., Picton L., Miller L., Strauch E.-M., DeBouver N. D., Pires A., Bera A. K., Halabiya S., Hammerson B., Yang W., Bernard S., Stewart L., Wilson I. A., Ruohola-Baker H., Schlessinger J., Lee S., Savvides S. N., Garcia K. C., Baker D., Design of protein-binding proteins from the target structure alone. Nature 605, 551–560 (2022). - PMC - PubMed
-
- Chen H., Simoska O., Lim K., Grattieri M., Yuan M., Dong F., Lee Y. S., Beaver K., Weliwatte S., Gaffney E. M., Minteer S. D., Fundamentals, applications, and future directions of Bioelectrocatalysis. Chem. Rev. 120, 12903–12993 (2020). - PubMed
-
- Sesterhenn F., Yang C., Bonet J., Cramer J. T., Wen X., Wang Y., Chiang C.-I., Abriata L. A., Kucharska I., Castoro G., Vollers S. S., Galloux M., Dheilly E., Rosset S., Corthésy P., Georgeon S., Villard M., Richard C.-A., Descamps D., Delgado T., Oricchio E., Rameix-Welti M.-A., Más V., Ervin S., Eléouët J.-F., Riffault S., Bates J. T., Julien J.-P., Li Y., Jardetzky T., Krey T., Correia B. E., De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science 368, (2020). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

