Augmenting L3MBTL2-induced condensates suppresses tumor growth in osteosarcoma
- PMID: 37992172
- PMCID: PMC10664996
- DOI: 10.1126/sciadv.adi0889
Augmenting L3MBTL2-induced condensates suppresses tumor growth in osteosarcoma
Abstract
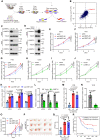
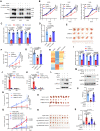
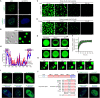
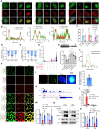
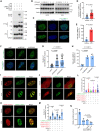
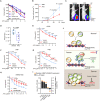
Osteosarcoma is a highly aggressive cancer and lacks effective therapeutic targets. We found that L3MBTL2 acts as a tumor suppressor by transcriptionally repressing IFIT2 in osteosarcoma. L3MBTL2 recruits the components of Polycomb repressive complex 1.6 to form condensates via both Pho-binding pockets and polybasic regions within carboxyl-terminal intrinsically disordered regions; the L3MBTL2-induced condensates are required for its tumor suppression. Multi-monoubiquitination of L3MBTL2 by UBE2O results in its proteasomal degradation, and the UBE2O/L3MBTL2 axis was crucial for osteosarcoma growth. There is a reverse correlation between L3MBTL2 and UBE2O in osteosarcoma tissues, and higher UBE2O and lower L3MBTL2 are associated with poorer prognosis in osteosarcoma. Pharmacological blockage of UBE2O by arsenic trioxide can enhance L3MBTL2-induced condensates and consequently suppress osteosarcoma growth. Our findings unveil a crucial biological function of L3MBTL2-induced condensates in mediating tumor suppression, proposing the UBE2O-L3MBTL2 axis as a potential cancer therapeutic target in osteosarcoma.
Figures


Similar articles
-
Ubiquitination in osteosarcoma: unveiling the impact on cell biology and therapeutic strategies.Cancer Biol Med. 2024 Oct 30;21(10):880-97. doi: 10.20892/j.issn.2095-3941.2024.0231. Cancer Biol Med. 2024. PMID: 39475222 Free PMC article. Review.
-
Ubiquitin-conjugating enzyme UBE2O regulates cellular clock function by promoting the degradation of the transcription factor BMAL1.J Biol Chem. 2018 Jul 20;293(29):11296-11309. doi: 10.1074/jbc.RA117.001432. Epub 2018 Jun 5. J Biol Chem. 2018. PMID: 29871923 Free PMC article.
-
UBE2O targets Mxi1 for ubiquitination and degradation to promote lung cancer progression and radioresistance.Cell Death Differ. 2021 Feb;28(2):671-684. doi: 10.1038/s41418-020-00616-8. Epub 2020 Sep 8. Cell Death Differ. 2021. PMID: 32901121 Free PMC article.
-
The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development.Cell Stem Cell. 2012 Sep 7;11(3):319-32. doi: 10.1016/j.stem.2012.06.002. Epub 2012 Jul 5. Cell Stem Cell. 2012. PMID: 22770845 Free PMC article.
-
Diverse roles of the E2/E3 hybrid enzyme UBE2O in the regulation of protein ubiquitination, cellular functions, and disease onset.FEBS J. 2019 Jun;286(11):2018-2034. doi: 10.1111/febs.14708. Epub 2018 Dec 4. FEBS J. 2019. PMID: 30468556 Review.
Cited by
-
Identification and functional characterization of T-cell exhaustion-associated lncRNA AL031775.1 in osteosarcoma: a novel therapeutic target.Front Immunol. 2025 Feb 24;16:1517971. doi: 10.3389/fimmu.2025.1517971. eCollection 2025. Front Immunol. 2025. PMID: 40066455 Free PMC article.
-
Long non-coding RNA AC133552.2: biomarker and therapeutic target in osteosarcoma via PANoptosis gene screening.Sci Rep. 2025 Mar 17;15(1):9180. doi: 10.1038/s41598-025-93167-2. Sci Rep. 2025. PMID: 40097576 Free PMC article.
-
PRKDC Induces Chemoresistance in Osteosarcoma by Recruiting GDE2 to Stabilize GNAS and Activate AKT.Cancer Res. 2024 Sep 4;84(17):2873-2887. doi: 10.1158/0008-5472.CAN-24-0163. Cancer Res. 2024. PMID: 38900943 Free PMC article.
-
The deubiquitinase USP44 enhances cisplatin chemosensitivity through stabilizing STUB1 to promote LRPPRC degradation in neuroblastoma.Neuro Oncol. 2025 Feb 10;27(2):492-507. doi: 10.1093/neuonc/noae175. Neuro Oncol. 2025. PMID: 39215663
-
Ubiquitination in osteosarcoma: unveiling the impact on cell biology and therapeutic strategies.Cancer Biol Med. 2024 Oct 30;21(10):880-97. doi: 10.20892/j.issn.2095-3941.2024.0231. Cancer Biol Med. 2024. PMID: 39475222 Free PMC article. Review.
References
-
- P. S. Meltzer, L. J. Helman, New horizons in the treatment of osteosarcoma. N. Engl. J. Med. 385, 2066–2076 (2021). - PubMed
-
- X. Chen, A. Bahrami, A. Pappo, J. Easton, J. Dalton, E. Hedlund, D. Ellison, S. Shurtleff, G. Wu, L. Wei, M. Parker, M. Rusch, P. Nagahawatte, J. Wu, S. Mao, K. Boggs, H. Mulder, D. Yergeau, C. Lu, L. Ding, M. Edmonson, C. Qu, J. Wang, Y. Li, F. Navid, N. C. Daw, E. R. Mardis, R. K. Wilson, J. R. Downing, J. Zhang, M. A. Dyer; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project , Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 7, 104–112 (2014). - PMC - PubMed
-
- L. C. Sayles, M. R. Breese, A. L. Koehne, S. G. Leung, A. G. Lee, H. Y. Liu, A. Spillinger, A. T. Shah, B. Tanasa, K. Straessler, F. K. Hazard, S. L. Spunt, N. Marina, G. E. Kim, S. J. Cho, R. S. Avedian, D. G. Mohler, M. O. Kim, S. G. DuBois, D. S. Hawkins, E. A. Sweet-Cordero, Genome-informed targeted therapy for osteosarcoma. Cancer Discov. 9, 46–63 (2019). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

