Bifunctional Small Molecules That Induce Nuclear Localization and Targeted Transcriptional Regulation
- PMID: 37992275
- PMCID: PMC10704550
- DOI: 10.1021/jacs.3c06179
Bifunctional Small Molecules That Induce Nuclear Localization and Targeted Transcriptional Regulation
Abstract
The aberrant localization of proteins in cells is a key factor in the development of various diseases, including cancer and neurodegenerative disease. To better understand and potentially manipulate protein localization for therapeutic purposes, we engineered bifunctional compounds that bind to proteins in separate cellular compartments. We show these compounds induce nuclear import of cytosolic cargoes, using nuclear-localized BRD4 as a "carrier" for co-import and nuclear trapping of cytosolic proteins. We use this system to calculate kinetic constants for passive diffusion across the nuclear pore and demonstrate single-cell heterogeneity in response to these bifunctional molecules with cells requiring high carrier to cargo expression for complete import. We also observe incorporation of cargo into BRD4-containing condensates. Proteins shown to be substrates for nuclear transport include oncogenic mutant nucleophosmin (NPM1c) and mutant PI3K catalytic subunit alpha (PIK3CAE545K), suggesting potential applications to cancer treatment. In addition, we demonstrate that chemically induced localization of BRD4 to cytosolic-localized DNA-binding proteins, namely, IRF1 with a nuclear export signal, induces target gene expression. These results suggest that induced localization of proteins with bifunctional molecules enables the rewiring of cell circuitry, with significant implications for disease therapy.
Conflict of interest statement
The authors declare the following competing financial interest(s): W.J.G. reports equity in Ampressa therapeutics; is on the scientific advisory board (SAB), and has received consulting fees from Esperion therapeutics, consulting fees from Belharra therapeutics, Boston Clinical Research Institute, Faze Medicines, ImmPACT-Bio, and nference. A.C. is a founder and SAB member of Photys therapeutics. M.M. reports consultant/advisory board/equity for DelveBio, Interline and Isabl; research funding from Janssen and Bayer Pharmaceuticals; patents licensed to LabCorp and Bayer. S.L.S. is a shareholder and serves on the Board of Directors of Kojin Therapeutics; is a shareholder and advises Jnana Therapeutics, Kisbee Therapeutics, Belharra Therapeutics, Magnet Biomedicine, Exo Therapeutics, Eikonizo Therapeutics, and Replay Bio; advises Vividion Therapeutics, Eisai Co., Ltd., Ono Pharma Foundation, F-Prime Capital Partners, and is a Novartis Faculty Scholar. All COI are outside the submitted work and all other authors report no COI.
Figures

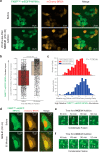
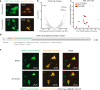

Update of
-
Bifunctional small molecules that induce nuclear localization and targeted transcriptional regulation.bioRxiv [Preprint]. 2023 Jul 7:2023.07.07.548101. doi: 10.1101/2023.07.07.548101. bioRxiv. 2023. Update in: J Am Chem Soc. 2023 Dec 6;145(48):26028-26037. doi: 10.1021/jacs.3c06179. PMID: 37461636 Free PMC article. Updated. Preprint.
Similar articles
-
Bifunctional small molecules that induce nuclear localization and targeted transcriptional regulation.bioRxiv [Preprint]. 2023 Jul 7:2023.07.07.548101. doi: 10.1101/2023.07.07.548101. bioRxiv. 2023. Update in: J Am Chem Soc. 2023 Dec 6;145(48):26028-26037. doi: 10.1021/jacs.3c06179. PMID: 37461636 Free PMC article. Updated. Preprint.
-
Molecular determinants of large cargo transport into the nucleus.Elife. 2020 Jul 21;9:e55963. doi: 10.7554/eLife.55963. Elife. 2020. PMID: 32692309 Free PMC article.
-
Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant-negative inhibitors.Curr Biol. 1998 Dec 17-31;8(25):1376-86. doi: 10.1016/s0960-9822(98)00018-9. Curr Biol. 1998. PMID: 9889100
-
Nucleocytoplasmic protein traffic and its significance to cell function.Genes Cells. 2000 Oct;5(10):777-87. doi: 10.1046/j.1365-2443.2000.00366.x. Genes Cells. 2000. PMID: 11029654 Review.
-
Therapeutic Dissolution of Aberrant Phases by Nuclear-Import Receptors.Trends Cell Biol. 2019 Apr;29(4):308-322. doi: 10.1016/j.tcb.2018.12.004. Epub 2019 Jan 16. Trends Cell Biol. 2019. PMID: 30660504 Free PMC article. Review.
Cited by
-
A biotin targeting chimera (BioTAC) system to map small molecule interactomes in situ.Nat Commun. 2023 Dec 4;14(1):8016. doi: 10.1038/s41467-023-43507-5. Nat Commun. 2023. PMID: 38049406 Free PMC article.
-
Temporal and Spatial Characterization of CUL3KLHL20-driven Targeted Degradation of BET family, BRD Proteins by the Macrocycle-based Degrader BTR2004.bioRxiv [Preprint]. 2024 Dec 7:2024.12.07.627262. doi: 10.1101/2024.12.07.627262. bioRxiv. 2024. Update in: ACS Chem Biol. 2025 Sep 2. doi: 10.1021/acschembio.5c00343. PMID: 39677683 Free PMC article. Updated. Preprint.
-
Covalent Proximity Inducers.Chem Rev. 2025 Jan 8;125(1):326-368. doi: 10.1021/acs.chemrev.4c00570. Epub 2024 Dec 18. Chem Rev. 2025. PMID: 39692621 Free PMC article. Review.
-
Regulated induced proximity targeting chimeras-RIPTACs-A heterobifunctional small molecule strategy for cancer selective therapies.Cell Chem Biol. 2024 Aug 15;31(8):1490-1502.e42. doi: 10.1016/j.chembiol.2024.07.005. Epub 2024 Aug 7. Cell Chem Biol. 2024. PMID: 39116881 Free PMC article.
-
Harnessing BET-Bromodomain Assisted Nuclear Import for Targeted Subcellular Localization and Enhanced Efficacy of Antisense Oligonucleotides.J Am Chem Soc. 2025 Aug 13;147(32):29478-29488. doi: 10.1021/jacs.5c09544. Epub 2025 Aug 4. J Am Chem Soc. 2025. PMID: 40758869 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

