Extracellular sodium regulates fibroblast growth factor 23 (FGF23) formation
- PMID: 37992803
- PMCID: PMC10770535
- DOI: 10.1016/j.jbc.2023.105480
Extracellular sodium regulates fibroblast growth factor 23 (FGF23) formation
Abstract
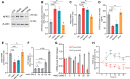
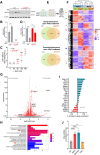
The bone-derived hormone fibroblast growth factor-23 (FGF23) has recently received much attention due to its association with chronic kidney disease and cardiovascular disease progression. Extracellular sodium concentration ([Na+]) plays a significant role in bone metabolism. Hyponatremia (lower serum [Na+]) has recently been shown to be independently associated with FGF23 levels in patients with chronic systolic heart failure. However, nothing is known about the direct impact of [Na+] on FGF23 production. Here, we show that an elevated [Na+] (+20 mM) suppressed FGF23 formation, whereas low [Na+] (-20 mM) increased FGF23 synthesis in the osteoblast-like cell lines UMR-106 and MC3T3-E1. Similar bidirectional changes in FGF23 abundance were observed when osmolality was altered by mannitol but not by urea, suggesting a role of tonicity in FGF23 formation. Moreover, these changes in FGF23 were inversely proportional to the expression of NFAT5 (nuclear factor of activated T cells-5), a transcription factor responsible for tonicity-mediated cellular adaptations. Furthermore, arginine vasopressin, which is often responsible for hyponatremia, did not affect FGF23 production. Next, we performed a comprehensive and unbiased RNA-seq analysis of UMR-106 cells exposed to low versus high [Na+], which revealed several novel genes involved in cellular adaptation to altered tonicity. Additional analysis of cells with Crisp-Cas9-mediated NFAT5 deletion indicated that NFAT5 controls numerous genes associated with FGF23 synthesis, thereby confirming its role in [Na+]-mediated FGF23 regulation. In line with these in vitro observations, we found that hyponatremia patients have higher FGF23 levels. Our results suggest that [Na+] is a critical regulator of FGF23 synthesis.
Keywords: FGF23; NFAT5; bone and kidney; extracellular sodium; hyponatremia.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no conflicts of interest with the contents of this article.
Figures



References
-
- White K.E., Evans W.E., O’Riordan J.L.H., Speer M.C., Econs M.J., Lorenz-Depiereux B., et al. Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat. Genet. 2000;26:345–348. - PubMed
-
- Liu S., Guo R., Simpson L.G., Xiao Z.S., Burnham C.E., Quarles L.D. Regulation of fibroblastic growth factor 23 expression but not degradation by PHEX. J. Biol. Chem. 2003;278:37419–37426. - PubMed
-
- Vervloet M. Renal and extrarenal effects of fibroblast growth factor 23. Nat. Rev. Nephrol. 2019;15:109–120. - PubMed

