Horizontal acquisition of a DNA ligase improves DNA damage tolerance in eukaryotes
- PMID: 37993452
- PMCID: PMC10665377
- DOI: 10.1038/s41467-023-43075-8
Horizontal acquisition of a DNA ligase improves DNA damage tolerance in eukaryotes
Abstract
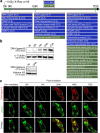
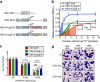
Bdelloid rotifers are part of the restricted circle of multicellular animals that can withstand a wide range of genotoxic stresses at any stage of their life cycle. In this study, bdelloid rotifer Adineta vaga is used as a model to decipher the molecular basis of their extreme tolerance. Proteomic analysis shows that a specific DNA ligase, different from those usually involved in DNA repair in eukaryotes, is strongly over-represented upon ionizing radiation. A phylogenetic analysis reveals its orthology to prokaryotic DNA ligase E, and its horizontal acquisition by bdelloid rotifers and plausibly other eukaryotes. The fungus Mortierella verticillata, having a single copy of this DNA Ligase E homolog, also exhibits an increased radiation tolerance with an over-expression of this DNA ligase E following X-ray exposure. We also provide evidence that A. vaga ligase E is a major contributor of DNA breaks ligation activity, which is a common step of all important DNA repair pathways. Consistently, its heterologous expression in human cell lines significantly improves their radio-tolerance. Overall, this study highlights the potential of horizontal gene transfers in eukaryotes, and their contribution to the adaptation to extreme conditions.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Friedberg, E. C. et al. DNA Repair and Mutagenesis, 2nd ed., (ASM Press, 2006).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

