Repeated Omicron exposures override ancestral SARS-CoV-2 immune imprinting
- PMID: 37993710
- PMCID: PMC10764275
- DOI: 10.1038/s41586-023-06753-7
Repeated Omicron exposures override ancestral SARS-CoV-2 immune imprinting
Abstract
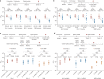
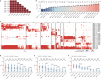
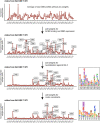
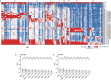
The continuing emergence of SARS-CoV-2 variants highlights the need to update COVID-19 vaccine compositions. However, immune imprinting induced by vaccination based on the ancestral (hereafter referred to as WT) strain would compromise the antibody response to Omicron-based boosters1-5. Vaccination strategies to counter immune imprinting are critically needed. Here we investigated the degree and dynamics of immune imprinting in mouse models and human cohorts, especially focusing on the role of repeated Omicron stimulation. In mice, the efficacy of single Omicron boosting is heavily limited when using variants that are antigenically distinct from WT-such as the XBB variant-and this concerning situation could be mitigated by a second Omicron booster. Similarly, in humans, repeated Omicron infections could alleviate WT vaccination-induced immune imprinting and generate broad neutralization responses in both plasma and nasal mucosa. Notably, deep mutational scanning-based epitope characterization of 781 receptor-binding domain (RBD)-targeting monoclonal antibodies isolated from repeated Omicron infection revealed that double Omicron exposure could induce a large proportion of matured Omicron-specific antibodies that have distinct RBD epitopes to WT-induced antibodies. Consequently, immune imprinting was largely mitigated, and the bias towards non-neutralizing epitopes observed in single Omicron exposures was restored. On the basis of the deep mutational scanning profiles, we identified evolution hotspots of XBB.1.5 RBD and demonstrated that these mutations could further boost the immune-evasion capability of XBB.1.5 while maintaining high ACE2-binding affinity. Our findings suggest that the WT component should be abandoned when updating COVID-19 vaccines, and individuals without prior Omicron exposure should receive two updated vaccine boosters.
© 2023. The Author(s).
Conflict of interest statement
X.S.X. and Y.C. are inventors on the provisional patent applications of BD series antibodies, which include BD55-5514 (SA55) and monoclonal antibodies from Omicron infection convalescents. X.S.X. and Y.C. are founders of Singlomics Biopharmaceuticals. The other authors declare no competing interests.
Figures










References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

