VGAE-MCTS: A New Molecular Generative Model Combining the Variational Graph Auto-Encoder and Monte Carlo Tree Search
- PMID: 37993764
- PMCID: PMC10716893
- DOI: 10.1021/acs.jcim.3c01220
VGAE-MCTS: A New Molecular Generative Model Combining the Variational Graph Auto-Encoder and Monte Carlo Tree Search
Abstract
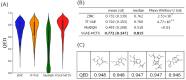
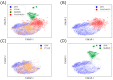
Molecular generation is crucial for advancing drug discovery, materials science, and chemical exploration. It expedites the search for new drug candidates, facilitates tailored material creation, and enhances our understanding of molecular diversity. By employing artificial intelligence techniques such as molecular generative models based on molecular graphs, researchers have tackled the challenge of identifying efficient molecules with desired properties. Here, we propose a new molecular generative model combining a graph-based deep neural network and a reinforcement learning technique. We evaluated the validity, novelty, and optimized physicochemical properties of the generated molecules. Importantly, the model explored uncharted regions of chemical space, allowing for the efficient discovery and design of new molecules. This innovative approach has considerable potential to revolutionize drug discovery, materials science, and chemical research for accelerating scientific innovation. By leveraging advanced techniques and exploring previously unexplored chemical spaces, this study offers promising prospects for the efficient discovery and design of new molecules in the field of drug development.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
De Novo Drug Design Using Transformer-Based Machine Translation and Reinforcement Learning of an Adaptive Monte Carlo Tree Search.Pharmaceuticals (Basel). 2024 Jan 27;17(2):161. doi: 10.3390/ph17020161. Pharmaceuticals (Basel). 2024. PMID: 38399376 Free PMC article.
-
Network-principled deep generative models for designing drug combinations as graph sets.Bioinformatics. 2020 Jul 1;36(Suppl_1):i445-i454. doi: 10.1093/bioinformatics/btaa317. Bioinformatics. 2020. PMID: 32657357 Free PMC article.
-
Enabling target-aware molecule generation to follow multi objectives with Pareto MCTS.Commun Biol. 2024 Sep 2;7(1):1074. doi: 10.1038/s42003-024-06746-w. Commun Biol. 2024. PMID: 39223327 Free PMC article.
-
Generative chemistry: drug discovery with deep learning generative models.J Mol Model. 2021 Feb 4;27(3):71. doi: 10.1007/s00894-021-04674-8. J Mol Model. 2021. PMID: 33543405 Free PMC article. Review.
-
Generative artificial intelligence empowers digital twins in drug discovery and clinical trials.Expert Opin Drug Discov. 2024 Jan-Jun;19(1):33-42. doi: 10.1080/17460441.2023.2273839. Epub 2024 Jan 8. Expert Opin Drug Discov. 2024. PMID: 37887266 Review.
Cited by
-
A 3D generation framework using diffusion model and reinforcement learning to generate multi-target compounds with desired properties.J Cheminform. 2025 Jun 4;17(1):93. doi: 10.1186/s13321-025-01035-y. J Cheminform. 2025. PMID: 40468393 Free PMC article.
-
DrugGym: A testbed for the economics of autonomous drug discovery.bioRxiv [Preprint]. 2024 Jun 2:2024.05.28.596296. doi: 10.1101/2024.05.28.596296. bioRxiv. 2024. PMID: 38854082 Free PMC article. Preprint.
-
Uncertainty quantification with graph neural networks for efficient molecular design.Nat Commun. 2025 Apr 5;16(1):3262. doi: 10.1038/s41467-025-58503-0. Nat Commun. 2025. PMID: 40188130 Free PMC article.
-
A systematic review of deep learning chemical language models in recent era.J Cheminform. 2024 Nov 18;16(1):129. doi: 10.1186/s13321-024-00916-y. J Cheminform. 2024. PMID: 39558376 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

