Exploring the molecular makeup of support cells in insect camera eyes
- PMID: 37993800
- PMCID: PMC10664524
- DOI: 10.1186/s12864-023-09804-5
Exploring the molecular makeup of support cells in insect camera eyes
Abstract
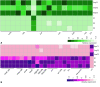
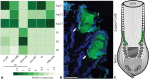
Animals typically have either compound eyes, or camera-type eyes, both of which have evolved repeatedly in the animal kingdom. Both eye types include two important kinds of cells: photoreceptor cells, which can be excited by light, and non-neuronal support cells (SupCs), which provide essential support to photoreceptors. At the molecular level deeply conserved genes that relate to the differentiation of photoreceptor cells have fueled a discussion on whether or not a shared evolutionary origin might be considered for this cell type. In contrast, only a handful of studies, primarily on the compound eyes of Drosophila melanogaster, have demonstrated molecular similarities in SupCs. D. melanogaster SupCs (Semper cells and primary pigment cells) are specialized eye glia that share several molecular similarities with certain vertebrate eye glia, including Müller glia. This led us to question if there could be conserved molecular signatures of SupCs, even in functionally different eyes such as the image-forming larval camera eyes of the sunburst diving beetle Thermonectus marmoratus. To investigate this possibility, we used an in-depth comparative whole-tissue transcriptomics approach. Specifically, we dissected the larval principal camera eyes into SupC- and retina-containing regions and generated the respective transcriptomes. Our analysis revealed several common features of SupCs including enrichment of genes that are important for glial function (e.g. gap junction proteins such as innexin 3), glycogen production (glycogenin), and energy metabolism (glutamine synthetase 1 and 2). To evaluate similarities, we compared our transcriptomes with those of fly (Semper cells) and vertebrate (Müller glia) eye glia as well as respective retinas. T. marmoratus SupCs were found to have distinct genetic overlap with both fly and vertebrate eye glia. These results suggest that T. marmoratus SupCs are a form of glia, and like photoreceptors, may be deeply conserved.
Keywords: Eye evolution; Gene regulatory network; Glia; Insects; Support cells; Transcriptomics.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Update of
-
Exploring the molecular makeup of support cells in insect camera eyes.bioRxiv [Preprint]. 2023 Jul 20:2023.07.19.549729. doi: 10.1101/2023.07.19.549729. bioRxiv. 2023. Update in: BMC Genomics. 2023 Nov 22;24(1):702. doi: 10.1186/s12864-023-09804-5. PMID: 37503285 Free PMC article. Updated. Preprint.
Similar articles
-
Exploring the molecular makeup of support cells in insect camera eyes.bioRxiv [Preprint]. 2023 Jul 20:2023.07.19.549729. doi: 10.1101/2023.07.19.549729. bioRxiv. 2023. Update in: BMC Genomics. 2023 Nov 22;24(1):702. doi: 10.1186/s12864-023-09804-5. PMID: 37503285 Free PMC article. Updated. Preprint.
-
Embryonic development of the larval eyes of the Sunburst Diving Beetle, Thermonectus marmoratus (Insecta: Dytiscidae): a morphological study.Evol Dev. 2016 Jul;18(4):216-28. doi: 10.1111/ede.12192. Evol Dev. 2016. PMID: 27402568
-
Multifunctional glial support by Semper cells in the Drosophila retina.PLoS Genet. 2017 May 31;13(5):e1006782. doi: 10.1371/journal.pgen.1006782. eCollection 2017 May. PLoS Genet. 2017. PMID: 28562601 Free PMC article.
-
Semper's cells in the insect compound eye: Insights into ocular form and function.Dev Biol. 2021 Nov;479:126-138. doi: 10.1016/j.ydbio.2021.07.015. Epub 2021 Jul 31. Dev Biol. 2021. PMID: 34343526 Free PMC article. Review.
-
How aquatic water-beetle larvae with small chambered eyes overcome challenges of hunting under water.J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2014 Nov;200(11):911-22. doi: 10.1007/s00359-014-0944-9. Epub 2014 Sep 27. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2014. PMID: 25261360 Review.
Cited by
-
Mitf, with Yki and STRIPAK-PP2A, is a key determinant of form and fate in the progenitor epithelium of the Drosophila eye.Eur J Cell Biol. 2024 Jun;103(2):151421. doi: 10.1016/j.ejcb.2024.151421. Epub 2024 May 15. Eur J Cell Biol. 2024. PMID: 38776620 Free PMC article.
-
Cataract induction in an arthropod reveals how lens crystallins contribute to the formation of biological glass.PLoS One. 2025 Jun 11;20(6):e0325229. doi: 10.1371/journal.pone.0325229. eCollection 2025. PLoS One. 2025. PMID: 40498792 Free PMC article.
-
Osmosis as nature's method for establishing optical alignment.Curr Biol. 2024 Apr 8;34(7):1569-1575.e3. doi: 10.1016/j.cub.2024.02.052. Epub 2024 Mar 20. Curr Biol. 2024. PMID: 38513653 Free PMC article.
References
-
- Land MF, Nilsson D-E. Animal Eyes. USA: Oxford University Press; 2012.
-
- Arendt D. Evolution of eyes and photoreceptor cell types. Int J Dev Biol. 2003;47:563–571. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

