Machine learning-derived identification of prognostic signature for improving prognosis and drug response in patients with ovarian cancer
- PMID: 37994489
- PMCID: PMC10805490
- DOI: 10.1111/jcmm.18021
Machine learning-derived identification of prognostic signature for improving prognosis and drug response in patients with ovarian cancer
Abstract
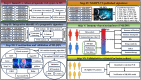
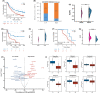
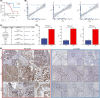
Clinical assessments relying on pathology classification demonstrate limited effectiveness in predicting clinical outcomes and providing optimal treatment for patients with ovarian cancer (OV). Consequently, there is an urgent requirement for an ideal biomarker to facilitate precision medicine. To address this issue, we selected 15 multicentre cohorts, comprising 12 OV cohorts and 3 immunotherapy cohorts. Initially, we identified a set of robust prognostic risk genes using data from the 12 OV cohorts. Subsequently, we employed a consensus cluster analysis to identify distinct clusters based on the expression profiles of the risk genes. Finally, a machine learning-derived prognostic signature (MLDPS) was developed based on differentially expressed genes and univariate Cox regression genes between the clusters by using 10 machine-learning algorithms (101 combinations). Patients with high MLDPS had unfavourable survival rates and have good prediction performance in all cohorts and in-house cohorts. The MLDPS exhibited robust and dramatically superior capability than 21 published signatures. Of note, low MLDIS have a positive prognostic impact on patients treated with anti-PD-1 immunotherapy by driving changes in the level of infiltration of immune cells. Additionally, patients suffering from OV with low MLDIS were more sensitive to immunotherapy. Meanwhile, patients with low MLDIS might benefit from chemotherapy, and 19 compounds that may be potential agents for patients with low MLDIS were identified. MLDIS presents an appealing instrument for the identification of patients at high/low risk. This could enhance the precision treatment, ultimately guiding the clinical management of OV.
Keywords: immunotherapy; machine learning; ovarian cancer; tumour microenvironment.
© 2023 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures





References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

