Comparative analysis of mitochondrion-related organelles in anaerobic amoebozoans
- PMID: 37994879
- PMCID: PMC10711303
- DOI: 10.1099/mgen.0.001143
Comparative analysis of mitochondrion-related organelles in anaerobic amoebozoans
Abstract
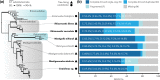
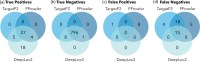
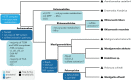
Archamoebae comprises free-living or endobiotic amoebiform protists that inhabit anaerobic or microaerophilic environments and possess mitochondrion-related organelles (MROs) adapted to function anaerobically. We compared in silico reconstructed MRO proteomes of eight species (six genera) and found that the common ancestor of Archamoebae possessed very few typical components of the protein translocation machinery, electron transport chain and tricarboxylic acid cycle. On the other hand, it contained a sulphate activation pathway and bacterial iron-sulphur (Fe-S) assembly system of MIS-type. The metabolic capacity of the MROs, however, varies markedly within this clade. The glycine cleavage system is widely conserved among Archamoebae, except in Entamoeba, probably owing to its role in catabolic function or one-carbon metabolism. MRO-based pyruvate metabolism was dispensed within subgroups Entamoebidae and Rhizomastixidae, whereas sulphate activation could have been lost in isolated cases of Rhizomastix libera, Mastigamoeba abducta and Endolimax sp. The MIS (Fe-S) assembly system was duplicated in the common ancestor of Mastigamoebidae and Pelomyxidae, and one of the copies took over Fe-S assembly in their MRO. In Entamoebidae and Rhizomastixidae, we hypothesize that Fe-S cluster assembly in both compartments may be facilitated by dual localization of the single system. We could not find evidence for changes in metabolic functions of the MRO in response to changes in habitat; it appears that such environmental drivers do not strongly affect MRO reduction in this group of eukaryotes.
Keywords: anaerobiosis; comparative genomics; mitochondrion-related organelles; reductive evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Gawryluk RMR, Chisholm KA, Pinto DM, Gray MW. Compositional complexity of the mitochondrial proteome of a unicellular eukaryote (Acanthamoeba castellanii, supergroup Amoebozoa) rivals that of animals, fungi, and plants. J Proteomics. 2014;109:400–416. doi: 10.1016/j.jprot.2014.07.005. - DOI - PubMed
-
- Atteia A, Adrait A, Brugière S, Tardif M, van Lis R, et al. A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol Biol Evol. 2009;26:1533–1548. doi: 10.1093/molbev/msp068. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

