Molecular basis of opioid receptor signaling
- PMID: 37995655
- PMCID: PMC10710086
- DOI: 10.1016/j.cell.2023.10.029
Molecular basis of opioid receptor signaling
Abstract
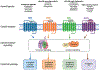
Opioids are used for pain management despite the side effects that contribute to the opioid crisis. The pursuit of non-addictive opioid analgesics remains unattained due to the unresolved intricacies of opioid actions, receptor signaling cascades, and neuronal plasticity. Advancements in structural, molecular, and computational tools illuminate the dynamic interplay between opioids and opioid receptors, as well as the molecular determinants of signaling pathways, which are potentially interlinked with pharmacological responses. Here, we review the molecular basis of opioid receptor signaling with a focus on the structures of opioid receptors bound to endogenous peptides or pharmacological agents. These insights unveil specific interactions that dictate ligand selectivity and likely their distinctive pharmacological profiles. Biochemical analysis further unveils molecular features governing opioid receptor signaling. Simultaneously, the synergy between computational biology and medicinal chemistry continues to expedite the discovery of novel chemotypes with the promise of yielding more efficacious and safer opioid compounds.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests B.L.R. is a co-founder of Epiodyne and is listed by the University of North Carolina Chapel Hill Medical School as an inventor on technologies and small molecules related to opioid receptors.
Figures
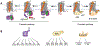




References
-
- Hippocrates, and Littré, T.E.m. (1853). Oeuvres complètes d’Hippocrate. Baillière, Paris.
-
- Serturner FWA (1805). Saure im Opium. Trommsdorffs J Pharm.
-
- Serturner FWA (1806). Darstellung der reinen Mohnsäure (Opiumsäure) nebst einer Chemischen Untersuchung des Opiums mit vorzüglicher Hinsicht auf einem darin neu entdeckten Stoff und die dahin gehörigen Bemerkungen. Neues J. Pharm 14, 47–93.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

