Programming of neural progenitors of the adult subependymal zone towards a glutamatergic neuron lineage by neurogenin 2
- PMID: 37995703
- PMCID: PMC10724369
- DOI: 10.1016/j.stemcr.2023.10.019
Programming of neural progenitors of the adult subependymal zone towards a glutamatergic neuron lineage by neurogenin 2
Abstract
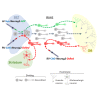
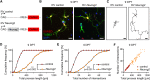
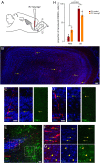
Although adult subependymal zone (SEZ) neural stem cells mostly generate GABAergic interneurons, a small progenitor population expresses the proneural gene Neurog2 and produces glutamatergic neurons. Here, we determined whether Neurog2 could respecify SEZ neural stem cells and their progeny toward a glutamatergic fate. Retrovirus-mediated expression of Neurog2 induced the glutamatergic lineage markers TBR2 and TBR1 in cultured SEZ progenitors, which differentiated into functional glutamatergic neurons. Likewise, Neurog2-transduced SEZ progenitors acquired glutamatergic neuron hallmarks in vivo. Intriguingly, they failed to migrate toward the olfactory bulb and instead differentiated within the SEZ or the adjacent striatum, where they received connections from local neurons, as indicated by rabies virus-mediated monosynaptic tracing. In contrast, lentivirus-mediated expression of Neurog2 failed to reprogram early SEZ neurons, which maintained GABAergic identity and migrated to the olfactory bulb. Our data show that NEUROG2 can program SEZ progenitors toward a glutamatergic identity but fails to reprogram their neuronal progeny.
Keywords: adult neurogenesis; adult subventricular zone; fate reprogramming; neural specification; neural stem cell; neurogenin 2; olfactory bulb; proneural gene.
Copyright © 2023. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

