Effective delivery of miR-511-3p with mannose-decorated exosomes with RNA nanoparticles confers protection against asthma
- PMID: 37996055
- PMCID: PMC10872989
- DOI: 10.1016/j.jconrel.2023.11.034
Effective delivery of miR-511-3p with mannose-decorated exosomes with RNA nanoparticles confers protection against asthma
Abstract
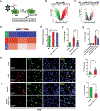
Our previous studies have shown that miR-511-3p treatment has a beneficial effect in alleviating allergic airway inflammation. Here, we sought to explore its therapeutic potential in animal models and gain a deeper understanding of its therapeutic value for asthma. miR-511-3p knockout mice (miR-511-3p-/-) were generated by CRISPR/Cas and showed exacerbated airway hyper-responsiveness and Th2-associated allergic airway inflammation compared with wild-type (WT) mice after exposed to cockroach allergen. RNA nanoparticles with mannose decorated EV-miR-511-3p were also created by loading miR-511-3p mimics into the mannose decorated EVs with engineered RNA nanoparticle PRNA-3WJ (Man-EV-miR-511-3p). Intra-tracheal inhalation of Man-EV-miR-511-3p, which could effectively penetrate the airway mucus barrier and deliver functional miR-511-3p to lung macrophages, successfully reversed the increased airway inflammation observed in miR-511-3p-/- mice. Through microarray analysis, complement C3 (C3) was identified as one of the major targets of miR-511-3p. C3 was increased in LPS-treated macrophages but decreased after miR-511-3p treatment. Consistent with these findings, C3 expression was elevated in the lung macrophages of an asthma mouse model but decreased in mice treated with miR-511-3p. Further experiments, including miRNA-mRNA pulldown and luciferase reporter assays, confirmed that miR-511-3p directly binds to C3 and activates the C3 gene. Thus, miR-511-3p represents a promising therapeutic target for asthma, and RNA nanotechnology reprogrammed EVs are efficient carriers for miRNA delivery for disease treatment.
Keywords: Asthma; Complement C3; Inflammation; Macrophage; miRNA.
Copyright © 2023. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors have declared that no conflict of interest exists.
Figures






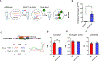
References
-
- Gelber LE, Seltzer LH, Bouzoukis JK, Pollart SM, Chapman MD, Platts-Mills TA. Sensitization and exposure to indoor allergens as risk factors for asthma among patients presenting to hospital. The American review of respiratory disease 1993; 147(3): 573–8. - PubMed
-
- Rosenstreich DL, Eggleston P, Kattan M, et al. The role of cockroach allergy and exposure to cockroach allergen in causing morbidity among inner-city children with asthma. The New England journal of medicine 1997; 336(19): 1356–63. - PubMed
-
- Chew GL, Perzanowski MS, Canfield SM, et al. Cockroach allergen levels and associations with cockroach-specific IgE. The Journal of allergy and clinical immunology 2008; 121(1): 240–5. - PubMed
-
- Kang B, Vellody D, Homburger H, Yunginger JW. Cockroach cause of allergic asthma. Its specificity and immunologic profile. The Journal of allergy and clinical immunology 1979; 63(2): 80–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

