Molecular EPISTOP, a comprehensive multi-omic analysis of blood from Tuberous Sclerosis Complex infants age birth to two years
- PMID: 37996417
- PMCID: PMC10667269
- DOI: 10.1038/s41467-023-42855-6
Molecular EPISTOP, a comprehensive multi-omic analysis of blood from Tuberous Sclerosis Complex infants age birth to two years
Abstract
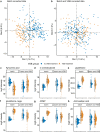
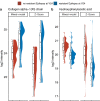
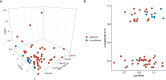
We present a comprehensive multi-omic analysis of the EPISTOP prospective clinical trial of early intervention with vigabatrin for pre-symptomatic epilepsy treatment in Tuberous Sclerosis Complex (TSC), in which 93 infants with TSC were followed from birth to age 2 years, seeking biomarkers of epilepsy development. Vigabatrin had profound effects on many metabolites, increasing serum deoxycytidine monophosphate (dCMP) levels 52-fold. Most serum proteins and metabolites, and blood RNA species showed significant change with age. Thirty-nine proteins, metabolites, and genes showed significant differences between age-matched control and TSC infants. Six also showed a progressive difference in expression between control, TSC without epilepsy, and TSC with epilepsy groups. A multivariate approach using enrollment samples identified multiple 3-variable predictors of epilepsy, with the best having a positive predictive value of 0.987. This rich dataset will enable further discovery and analysis of developmental effects, and associations with seizure development in TSC.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures