Hi-C metagenome sequencing reveals soil phage-host interactions
- PMID: 37996432
- PMCID: PMC10667309
- DOI: 10.1038/s41467-023-42967-z
Hi-C metagenome sequencing reveals soil phage-host interactions
Abstract
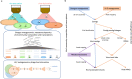
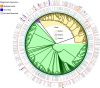
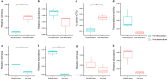
Bacteriophages are abundant in soils. However, the majority are uncharacterized, and their hosts are unknown. Here, we apply high-throughput chromosome conformation capture (Hi-C) to directly capture phage-host relationships. Some hosts have high centralities in bacterial community co-occurrence networks, suggesting phage infections have an important impact on the soil bacterial community interactions. We observe increased average viral copies per host (VPH) and decreased viral transcriptional activity following a two-week soil-drying incubation, indicating an increase in lysogenic infections. Soil drying also alters the observed phage host range. A significant negative correlation between VPH and host abundance prior to drying indicates more lytic infections result in more host death and inversely influence host abundance. This study provides empirical evidence of phage-mediated bacterial population dynamics in soil by directly capturing specific phage-host interactions.
© 2023. Battelle Memorial Institute.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Jansson, J. K. & Wu, R. Soil viral diversity, ecology and climate change. Nat. Rev. Microbiol. 1–16 (2022). - PubMed
-
- Schimel JP. Life in dry soils: effects of drought on soil microbial communities and processes. Annu Rev. Ecol. Evol. Syst. 2018;49:409–432. doi: 10.1146/annurev-ecolsys-110617-062614. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

