Complete chloroplast genomes of six neotropical palm species, structural comparison, and evolutionary dynamic patterns
- PMID: 37996522
- PMCID: PMC10667357
- DOI: 10.1038/s41598-023-44631-4
Complete chloroplast genomes of six neotropical palm species, structural comparison, and evolutionary dynamic patterns
Abstract
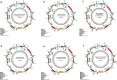
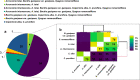
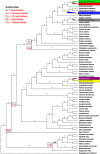
The Arecaceae family has a worldwide distribution, especially in tropical and subtropical regions. We sequenced the chloroplast genomes of Acrocomia intumescens and A. totai, widely used in the food and energy industries; Bactris gasipaes, important for palm heart; Copernicia alba and C. prunifera, worldwide known for wax utilization; and Syagrus romanzoffiana, of great ornamental potential. Copernicia spp. showed the largest chloroplast genomes (C. prunifera: 157,323 bp and C. alba: 157,192 bp), while S. romanzoffiana and B. gasipaes var. gasipaes presented the smallest (155,078 bp and 155,604 bp). Structurally, great synteny was detected among palms. Conservation was also observed in the distribution of single sequence repeats (SSR). Copernicia spp. presented less dispersed repeats, without occurrence in the small single copy (SSC). All RNA editing sites were C (cytidine) to U (uridine) conversions. Overall, closely phylogenetically related species shared more sites. Almost all nodes of the phylogenetic analysis showed a posterior probability (PP) of 1.0, reaffirming the close relationship between Acrocomia species. These results elucidate the conservation among palm chloroplast genomes, but point to subtle structural changes, providing support for the evolutionary dynamics of the Arecaceae family.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Baker, W. J. & Couvreur, T. L. P. Global biogeography and diversification of palms sheds light on the evolution of tropical lineages. II. Diversification history and origin of regional assemblages. J. Biogeogr.40, 286–298. 10.1111/j.1365-2699.2012.02794.x (2013).
-
- Muscarella R, et al. The global abundance of tree palms. Glob. Ecol. Biogeogr. 2020;29:1495–1514. doi: 10.1111/GEB.13123. - DOI
-
- Balslev H, et al. Species diversity and growth forms in tropical American palm Communities. Bot. Rev. 2011;77:381–425. doi: 10.1007/s12229-011-9084-x. - DOI
-
- Henderson A, Galeano G, Bernal R. Field guide to the palms of the Americas. Syst. Bot. 1996;21:258–259. doi: 10.2307/2419756. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

