Intra-promoter switch of transcription initiation sites in proliferation signaling-dependent RNA metabolism
- PMID: 37996663
- PMCID: PMC10716046
- DOI: 10.1038/s41594-023-01156-8
Intra-promoter switch of transcription initiation sites in proliferation signaling-dependent RNA metabolism
Abstract
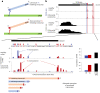
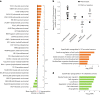
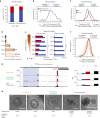
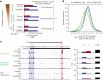
Global changes in transcriptional regulation and RNA metabolism are crucial features of cancer development. However, little is known about the role of the core promoter in defining transcript identity and post-transcriptional fates, a potentially crucial layer of transcriptional regulation in cancer. In this study, we use CAGE-seq analysis to uncover widespread use of dual-initiation promoters in which non-canonical, first-base-cytosine (C) transcription initiation occurs alongside first-base-purine initiation across 59 human cancers and healthy tissues. C-initiation is often followed by a 5' terminal oligopyrimidine (5'TOP) sequence, dramatically increasing the range of genes potentially subjected to 5'TOP-associated post-transcriptional regulation. We show selective, dynamic switching between purine and C-initiation site usage, indicating transcription initiation-level regulation in cancers. We additionally detail global metabolic changes in C-initiation transcripts that mark differentiation status, proliferative capacity, radiosensitivity, and response to irradiation and to PI3K-Akt-mTOR and DNA damage pathway-targeted radiosensitization therapies in colorectal cancer organoids and cancer cell lines and tissues.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

