Assembly and analysis of the complete mitochondrial genome of Forsythia suspensa (Thunb.) Vahl
- PMID: 37996801
- PMCID: PMC10666317
- DOI: 10.1186/s12864-023-09821-4
Assembly and analysis of the complete mitochondrial genome of Forsythia suspensa (Thunb.) Vahl
Abstract
Background: Forsythia suspensa (Thunb.) Vahl is a valuable ornamental and medicinal plant. Although the nuclear and chloroplast genomes of F. suspensa have been published, its complete mitochondrial genome sequence has yet to be reported. In this study, the genomic DNA of F. suspensa yellowish leaf material was extracted, sequenced by using a mixture of Illumina Novaseq6000 short reads and Oxford Nanopore PromethION long reads, and the sequencing data were assembled and annotated.
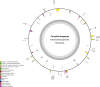
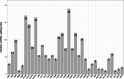
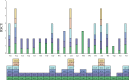
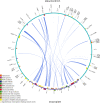
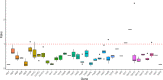
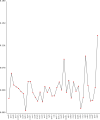
Result: The F. suspensa mitochondrial genome was obtained in the length of 535,692 bp with a circular structure, and the GC content was 44.90%. The genome contains 60 genes, including 36 protein-coding genes, 21 tRNA genes, and three rRNA genes. We further analyzed RNA editing of the protein-coding genes, relative synonymous codon usage, and sequence repeats based on the genomic data. There were 25 homologous sequences between F. suspensa mitochondria and chloroplast genome, which involved the transfer of 8 mitochondrial genes, and 9473 homologous sequences between mitochondrial and nuclear genomes. Analysis of the nucleic acid substitution rate, nucleic acid diversity, and collinearity of protein-coding genes of the F. suspensa mitochondrial genome revealed that the majority of genes may have undergone purifying selection, exhibiting a slower rate of evolution and a relatively conserved structure. Analysis of the phylogenetic relationships among different species revealed that F. suspensa was most closely related to Olea europaea subsp. Europaea.
Conclusion: In this study, we sequenced, assembled, and annotated a high-quality F. suspensa mitochondrial genome. The results of this study will enrich the mitochondrial genome data of Forsythia, lay a foundation for the phylogenetic development of Forsythia, and promote the evolutionary analysis of Oleaceae species.
Keywords: Bioinformatics analysis; Forsythia suspensa (Thunb.) Vahl; Mitochondrial genome; Phylogenetic evolution.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
The Complete Chloroplast Genome Sequences of the Medicinal Plant Forsythia suspensa (Oleaceae).Int J Mol Sci. 2017 Oct 31;18(11):2288. doi: 10.3390/ijms18112288. Int J Mol Sci. 2017. PMID: 29088105 Free PMC article.
-
[Intraspecific variation of Forsythia suspensa chloroplast genome].Zhongguo Zhong Yao Za Zhi. 2025 Apr;50(8):2108-2115. doi: 10.19540/j.cnki.cjcmm.20250114.103. Zhongguo Zhong Yao Za Zhi. 2025. PMID: 40461220 Chinese.
-
Assembly and comparative analysis of the complete mitochondrial genome of Fritillaria ussuriensis Maxim. (Liliales: Liliaceae), an endangered medicinal plant.BMC Genomics. 2024 Aug 8;25(1):773. doi: 10.1186/s12864-024-10680-w. BMC Genomics. 2024. PMID: 39118028 Free PMC article.
-
Phytochemistry, pharmacology, quality control and future research of Forsythia suspensa (Thunb.) Vahl: A review.J Ethnopharmacol. 2018 Jan 10;210:318-339. doi: 10.1016/j.jep.2017.08.040. Epub 2017 Sep 5. J Ethnopharmacol. 2018. PMID: 28887216 Review.
-
UPLC/Q-TOF MS Screening and Identification of Antibacterial Compounds in Forsythia suspensa (Thunb.) Vahl Leaves.Front Pharmacol. 2022 Jan 28;12:704260. doi: 10.3389/fphar.2021.704260. eCollection 2021. Front Pharmacol. 2022. PMID: 35153732 Free PMC article. Review.
Cited by
-
Comparative analysis of the complete mitochondrial genomes of Firmiana danxiaensis and F. kwangsiensis (Malvaceae), two endangered Firmiana species in China.Planta. 2025 Apr 9;261(5):107. doi: 10.1007/s00425-025-04685-2. Planta. 2025. PMID: 40205193
-
Complete mitochondrial genome of Polygonatum cyrtonema Hua reveals variation diversity and evolutionary trends.BMC Plant Biol. 2025 Aug 21;25(1):1109. doi: 10.1186/s12870-025-07074-9. BMC Plant Biol. 2025. PMID: 40841885
-
The Complete Mitochondrial Genome Sequence of Eimeria kongi (Apicomplexa: Coccidia).Life (Basel). 2024 May 29;14(6):699. doi: 10.3390/life14060699. Life (Basel). 2024. PMID: 38929682 Free PMC article.
-
De novo assembly and comparative analysis of cherry (Prunus subgenus Cerasus) mitogenomes.Front Plant Sci. 2025 Mar 24;16:1568698. doi: 10.3389/fpls.2025.1568698. eCollection 2025. Front Plant Sci. 2025. PMID: 40196431 Free PMC article.
-
Tracing post-domestication historical events and screening pre-breeding germplasm from large gene pools in wheat in the absence of phenotype data.Theor Appl Genet. 2024 Sep 28;137(10):237. doi: 10.1007/s00122-024-04738-2. Theor Appl Genet. 2024. PMID: 39340687
References
-
- State Pharmacopoeia Commission of the PRC . Pharmacopoeia of The people’s Republic of China. Volume I, Beijing: China Medical Science and Technology Press; 2020. p. 177.
-
- Fu ZZ, Li YH, Zhang KM, Li Y. Molecular data and ecological niche modeling reveal population dynamics of widespread shrub Forsythia suspensa (Oleaceae) in China’s warm-temperate zone in response to climate change during the Pleistocene. BMC Evol Biol. 2014;14:114. doi: 10.1186/1471-2148-14-114. - DOI - PMC - PubMed
-
- Spencer CH, Barrett LK, Jesson AM. Baker. The evolution and function of Stylar polymorphisms in flowering plants. Ann Botany. 2000;85.
-
- Hu K, Guan WJ, Bi Y, Zhang W, Li L, Zhang B, Liu Q, Song Y, Li X, Duan Z, Zheng Q, Yang Z, Liang J, Han M, Ruan L, Wu C, Zhang Y, Jia ZH, Zhong NS. Efficacy and safety of Lianhuaqingwen capsules, a repurposed Chinese herb, in patients with coronavirus Disease 2019: a multicenter, prospective, randomized controlled trial. Phytomedicine. 2021;85:153242. doi: 10.1016/j.phymed.2020.153242. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

