Detection of monkeypox virus using helicase dependent amplification and recombinase polymerase amplification combined with lateral flow test
- PMID: 37996921
- PMCID: PMC10668421
- DOI: 10.1186/s12985-023-02223-8
Detection of monkeypox virus using helicase dependent amplification and recombinase polymerase amplification combined with lateral flow test
Abstract
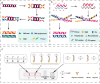
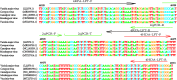
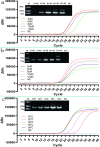
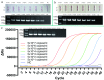
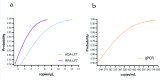
The monkeypox virus (MPXV) is a zoonotic DNA virus that belongs to the poxvirus family. Conventional laboratory methods for detecting MPXV are complex and expensive, making them unsuitable for detecting the virus in regions with limited resources. In this study, we using the Helicase dependent amplification (HDA) method and the Recombinase polymerase amplification (RPA) technique in combination with the lateral flow test (LFT), together with a self-designed qPCR technique for the detection of the MPXV specific conserved fragment F3L, to compare the sensitivity and specificity of the three assays. By analyzing the sensitivity detection results using Probit, it can be seen that the limit of detection (LOD) of the HDA-LFT detection target is 9.86 copies/µL (95% confidence interval, CI 7.52 copies/µL lower bound), the RPA-LFT detection target is 6.97 copies/µL (95% CI 3.90 copies/µL lower bound), and the qPCR detection target is 479.24 copies/mL (95% CI 273.81 copies/mL lower bound). The specificity test results showed that the specificity of the three methods mentioned above was higher than 90% in detecting pseudoviruses of the same genus of MPXV. The simple, highly sensitive, and specific MPXV assay developed in this study is anticipated to provide a solid foundation for future applications in the early screening, diagnosis, and evaluation of the efficacy of MPXV. This is the first time the HDA-LFT assay has been utilized to detect MPXV infection.
Keywords: HDA; INAAT; LFT; Monkeypox; RPA; qPCR.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Mpox disease, diagnosis, and point of care platforms.Bioeng Transl Med. 2025 Jan 2;10(3):e10733. doi: 10.1002/btm2.10733. eCollection 2025 May. Bioeng Transl Med. 2025. PMID: 40385539 Free PMC article. Review.
-
Development and Characterization of Recombinase-Based Isothermal Amplification Assays (RPA/RAA) for the Rapid Detection of Monkeypox Virus.Viruses. 2022 Sep 23;14(10):2112. doi: 10.3390/v14102112. Viruses. 2022. PMID: 36298667 Free PMC article.
-
Recombinase polymerase amplification assay for rapid detection of Monkeypox virus.Diagn Microbiol Infect Dis. 2019 Sep;95(1):41-45. doi: 10.1016/j.diagmicrobio.2019.03.015. Epub 2019 Apr 11. Diagn Microbiol Infect Dis. 2019. PMID: 31126795 Free PMC article.
-
Evaluation and clinical validation of monkeypox (mpox) virus real-time PCR assays.J Clin Virol. 2023 Feb;159:105373. doi: 10.1016/j.jcv.2022.105373. Epub 2022 Dec 23. J Clin Virol. 2023. PMID: 36603329 Free PMC article.
-
The current status and future prospects of CRISPR-based detection of monkeypox virus: A review.Anal Chim Acta. 2025 Jan 22;1336:343295. doi: 10.1016/j.aca.2024.343295. Epub 2024 Oct 1. Anal Chim Acta. 2025. PMID: 39788645 Review.
Cited by
-
Mpox disease, diagnosis, and point of care platforms.Bioeng Transl Med. 2025 Jan 2;10(3):e10733. doi: 10.1002/btm2.10733. eCollection 2025 May. Bioeng Transl Med. 2025. PMID: 40385539 Free PMC article. Review.
-
Lateral flow biosensor development for the visual identification of H1N1 virus based on primer extension nucleic acid isothermal amplification and M13mp18 single-stranded DNA.Mikrochim Acta. 2025 Feb 18;192(3):171. doi: 10.1007/s00604-025-07031-1. Mikrochim Acta. 2025. PMID: 39966254
-
Advances in Virus Detection Techniques Based on Recombinant Polymerase Amplification.Molecules. 2024 Oct 21;29(20):4972. doi: 10.3390/molecules29204972. Molecules. 2024. PMID: 39459340 Free PMC article. Review.
References
-
- Nolen LD, Osadebe L, Katomba J, Likofata J, Mukadi D, Monroe B, Doty J, Malekani J, Kabamba J, Bomponda PL. Introduction of monkeypox into a community and household: risk factors and zoonotic reservoirs in the Democratic Republic of the Congo. Am J Trop Med Hyg. 2015;93:410. doi: 10.4269/ajtmh.15-0168. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

